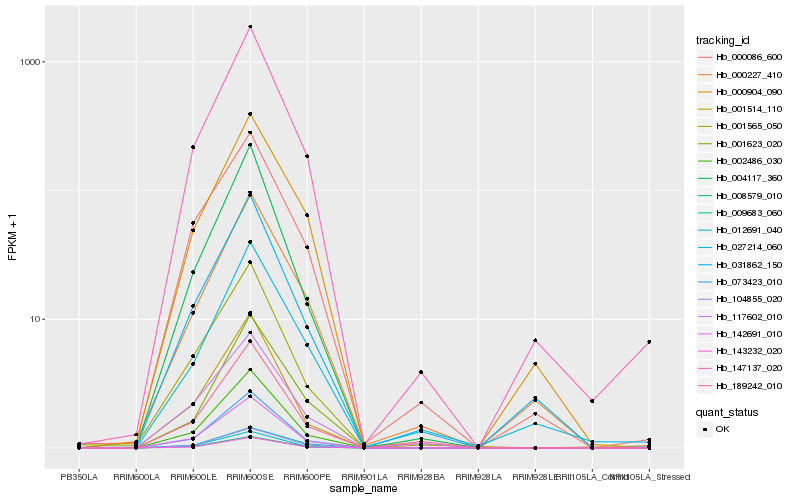
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147137_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 2 |
Hb_008579_010 |
0.0610007439 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 3 |
Hb_002486_030 |
0.0624899457 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 4 |
Hb_142691_010 |
0.0626691785 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 5 |
Hb_073423_010 |
0.0660639757 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_104855_020 |
0.067722271 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_031862_150 |
0.0688093087 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 8 |
Hb_012691_040 |
0.0720939763 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Fragaria vesca subsp. vesca] |
| 9 |
Hb_143232_020 |
0.0750956992 |
- |
- |
hypothetical protein JCGZ_20467 [Jatropha curcas] |
| 10 |
Hb_000227_410 |
0.0787001839 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 11 |
Hb_000904_090 |
0.0804858633 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 12 |
Hb_001565_050 |
0.0825454253 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Eucalyptus grandis] |
| 13 |
Hb_004117_360 |
0.0829534838 |
- |
- |
hypothetical protein JCGZ_00367 [Jatropha curcas] |
| 14 |
Hb_000086_600 |
0.0836132723 |
- |
- |
- |
| 15 |
Hb_001514_110 |
0.0861393235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_027214_060 |
0.0880110007 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 17 |
Hb_117602_010 |
0.0931616518 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 18 |
Hb_009683_060 |
0.0937403603 |
- |
- |
- |
| 19 |
Hb_189242_010 |
0.097796099 |
- |
- |
hypothetical protein B456_013G0075002, partial [Gossypium raimondii] |
| 20 |
Hb_001623_020 |
0.0978066902 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |