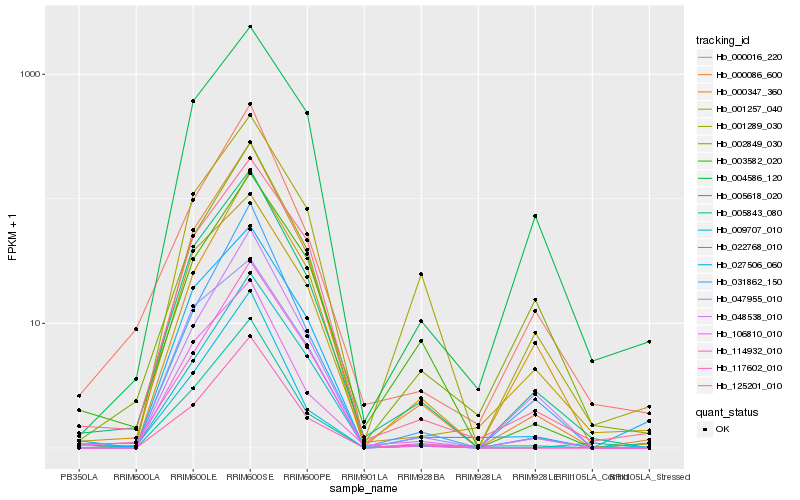
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005618_020 |
0.0 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 2 |
Hb_000086_600 |
0.0625638071 |
- |
- |
- |
| 3 |
Hb_106810_010 |
0.0801188432 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 4 |
Hb_125201_010 |
0.0820417289 |
- |
- |
hypothetical protein POPTR_0004s03690g [Populus trichocarpa] |
| 5 |
Hb_004586_120 |
0.0868004022 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 6 |
Hb_117602_010 |
0.0953561021 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 7 |
Hb_000016_220 |
0.0961454526 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 8 |
Hb_001257_040 |
0.0997969909 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 9 |
Hb_027506_060 |
0.1015988009 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 10 |
Hb_022768_010 |
0.1037029218 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 11 |
Hb_009707_010 |
0.1037643827 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 12 |
Hb_005843_080 |
0.1050404911 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_047955_010 |
0.1058473917 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 14 |
Hb_048538_010 |
0.1061483718 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Prunus mume] |
| 15 |
Hb_000347_360 |
0.1092442694 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002849_030 |
0.1092793738 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 17 |
Hb_031862_150 |
0.1098966751 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 18 |
Hb_114932_010 |
0.110081702 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_003582_020 |
0.1101205774 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Eucalyptus grandis] |
| 20 |
Hb_001289_030 |
0.1109897297 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |