| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
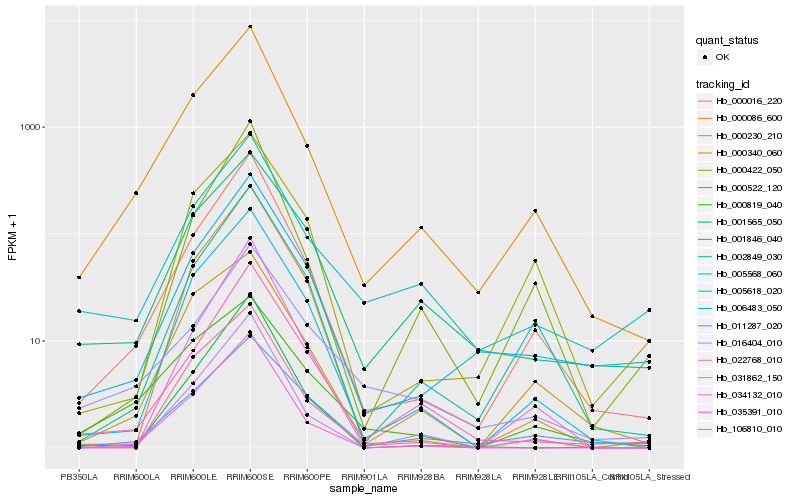
Hb_000230_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 2 |
Hb_000016_220 |
0.0755755171 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 3 |
Hb_005618_020 |
0.1149033678 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 4 |
Hb_022768_010 |
0.1276057864 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 5 |
Hb_002849_030 |
0.1284929947 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 6 |
Hb_034132_010 |
0.1295021723 |
- |
- |
hypothetical protein B456_007G015400 [Gossypium raimondii] |
| 7 |
Hb_106810_010 |
0.1309149707 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 8 |
Hb_035391_010 |
0.135735614 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 9 |
Hb_000340_060 |
0.1357679627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006483_050 |
0.1358276217 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 11 |
Hb_000819_040 |
0.1370463324 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000522_120 |
0.1373756063 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 13 |
Hb_011287_020 |
0.1407820892 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_005568_060 |
0.1414319125 |
- |
- |
PREDICTED: mitogen-activated protein kinase 3 [Jatropha curcas] |
| 15 |
Hb_001846_040 |
0.1431811891 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000086_600 |
0.1469125321 |
- |
- |
- |
| 17 |
Hb_001565_050 |
0.1479419053 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Eucalyptus grandis] |
| 18 |
Hb_016404_010 |
0.1486029899 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 19 |
Hb_031862_150 |
0.15000173 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 20 |
Hb_000422_050 |
0.1519048974 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |