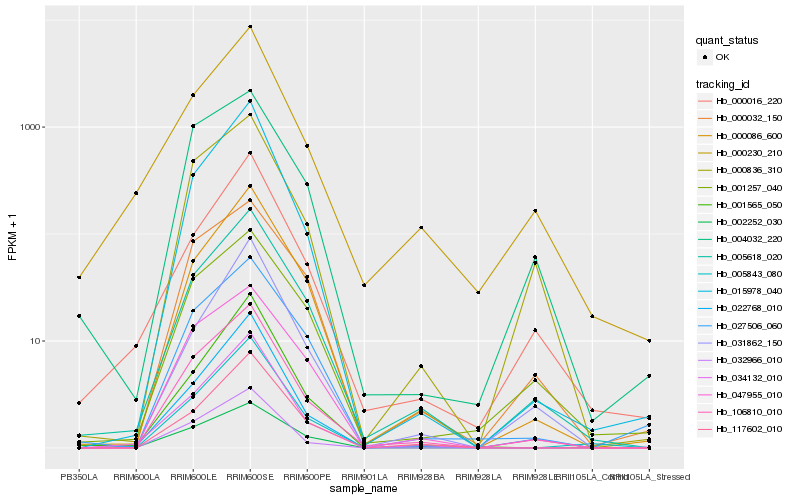
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106810_010 |
0.0 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_005618_020 |
0.0801188432 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 3 |
Hb_000836_310 |
0.0878441008 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 30 [Jatropha curcas] |
| 4 |
Hb_000086_600 |
0.0913615229 |
- |
- |
- |
| 5 |
Hb_015978_040 |
0.0918256224 |
- |
- |
PREDICTED: uncharacterized protein LOC105642609 [Jatropha curcas] |
| 6 |
Hb_022768_010 |
0.0947792296 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 7 |
Hb_032966_010 |
0.0954700419 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 8 |
Hb_047955_010 |
0.1026969431 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 9 |
Hb_034132_010 |
0.1042950431 |
- |
- |
hypothetical protein B456_007G015400 [Gossypium raimondii] |
| 10 |
Hb_005843_080 |
0.1076487704 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_117602_010 |
0.110196152 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 12 |
Hb_000032_150 |
0.114768404 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1691430 [Ricinus communis] |
| 13 |
Hb_027506_060 |
0.1173501736 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 14 |
Hb_004032_220 |
0.123309201 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 15 |
Hb_002252_030 |
0.1234245833 |
- |
- |
hypothetical protein JCGZ_06771 [Jatropha curcas] |
| 16 |
Hb_001257_040 |
0.1236994817 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 17 |
Hb_001565_050 |
0.1258913546 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Eucalyptus grandis] |
| 18 |
Hb_031862_150 |
0.1272665638 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 19 |
Hb_000016_220 |
0.1301010827 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 20 |
Hb_000230_210 |
0.1309149707 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |