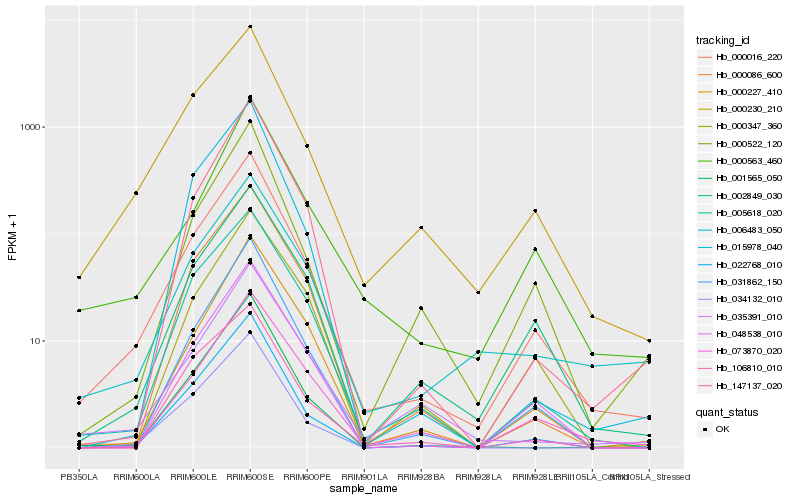
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_220 |
0.0 |
transcription factor |
TF Family: AP2 |
CRT/DRE binding factor 1 [Hevea brasiliensis] |
| 2 |
Hb_000230_210 |
0.0755755171 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 3 |
Hb_022768_010 |
0.0921643497 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 4 |
Hb_005618_020 |
0.0961454526 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 5 |
Hb_002849_030 |
0.096626701 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 6 |
Hb_031862_150 |
0.096673432 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 7 |
Hb_048538_010 |
0.108632657 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Prunus mume] |
| 8 |
Hb_000522_120 |
0.1117312839 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 9 |
Hb_001565_050 |
0.1131726888 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Eucalyptus grandis] |
| 10 |
Hb_073870_020 |
0.1132357443 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 11 |
Hb_000086_600 |
0.1142955131 |
- |
- |
- |
| 12 |
Hb_000563_460 |
0.116950502 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 13 |
Hb_000227_410 |
0.1171860999 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 14 |
Hb_006483_050 |
0.1172651024 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 15 |
Hb_034132_010 |
0.1172730842 |
- |
- |
hypothetical protein B456_007G015400 [Gossypium raimondii] |
| 16 |
Hb_147137_020 |
0.1188565458 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 17 |
Hb_035391_010 |
0.1245041831 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 18 |
Hb_000347_360 |
0.1248746011 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_106810_010 |
0.1301010827 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 20 |
Hb_015978_040 |
0.1307845673 |
- |
- |
PREDICTED: uncharacterized protein LOC105642609 [Jatropha curcas] |