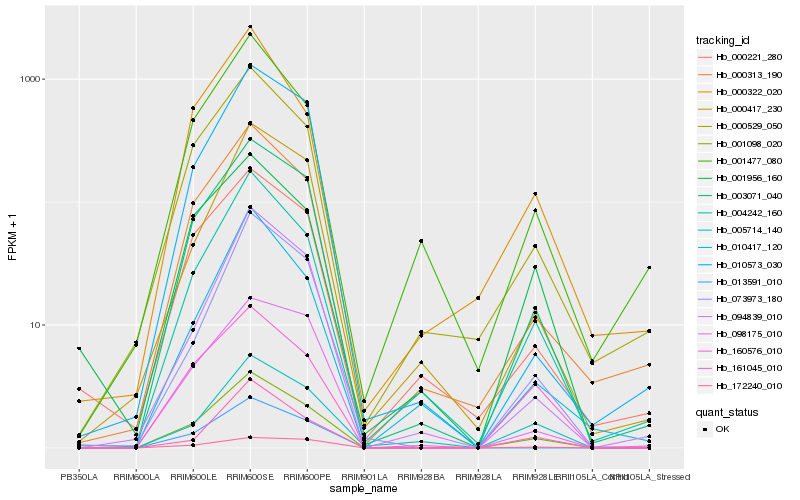
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001098_020 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 2 |
Hb_003071_040 |
0.0673500839 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 3 |
Hb_004242_160 |
0.0884254462 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000313_190 |
0.1083022867 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 5 |
Hb_000529_050 |
0.1088697592 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 6 |
Hb_073973_180 |
0.1136992985 |
- |
- |
PREDICTED: WAT1-related protein At4g08290 isoform X1 [Populus euphratica] |
| 7 |
Hb_094839_010 |
0.1228253755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_160576_010 |
0.1288550952 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000417_230 |
0.1314228173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010417_120 |
0.1324129876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_098175_010 |
0.1324639644 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 12 |
Hb_000221_280 |
0.1338652233 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 13 |
Hb_161045_010 |
0.1339581134 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 14 |
Hb_001477_080 |
0.1347430243 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 15 |
Hb_001956_160 |
0.1375207186 |
- |
- |
hypothetical protein L484_009438 [Morus notabilis] |
| 16 |
Hb_010573_030 |
0.137744438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000322_020 |
0.1379836116 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 18 |
Hb_013591_010 |
0.1385789266 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 19 |
Hb_005714_140 |
0.1388085894 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_172240_010 |
0.138962427 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |