| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
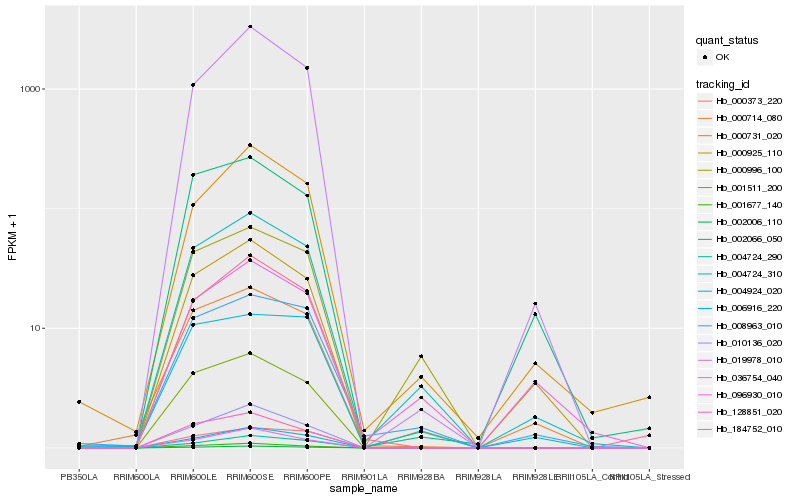
Hb_004724_310 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 2 |
Hb_006916_220 |
0.0717683072 |
- |
- |
hypothetical protein JCGZ_20099 [Jatropha curcas] |
| 3 |
Hb_000996_100 |
0.0856877785 |
- |
- |
Nucleotide binding, putative [Theobroma cacao] |
| 4 |
Hb_008963_010 |
0.0862869881 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 5 |
Hb_000925_110 |
0.0885338145 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 6 |
Hb_000731_020 |
0.0986441264 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 7 |
Hb_000714_080 |
0.102864211 |
transcription factor |
TF Family: Trihelix |
GT-2 factor [Glycine max] |
| 8 |
Hb_001511_200 |
0.1050814752 |
- |
- |
Embryonic abundant protein, putative [Ricinus communis] |
| 9 |
Hb_002006_110 |
0.1062804678 |
transcription factor |
TF Family: LFY |
LEAFY-like protein [Pistacia chinensis] |
| 10 |
Hb_010136_020 |
0.1063746632 |
- |
- |
stress-induced receptor-like kinase [Medicago truncatula] |
| 11 |
Hb_019978_010 |
0.1067845988 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001677_140 |
0.1092292262 |
- |
- |
PREDICTED: uncharacterized protein LOC102622520 isoform X1 [Citrus sinensis] |
| 13 |
Hb_184752_010 |
0.1109489799 |
- |
- |
PREDICTED: uncharacterized protein LOC105642593 [Jatropha curcas] |
| 14 |
Hb_096930_010 |
0.114596667 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 15 |
Hb_004724_290 |
0.1151522479 |
transcription factor |
TF Family: NAC |
- |
| 16 |
Hb_128851_020 |
0.1161303068 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_004924_020 |
0.1167625543 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_036754_040 |
0.1178162421 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_002066_050 |
0.12281411 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 20 |
Hb_000373_220 |
0.1242033289 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |