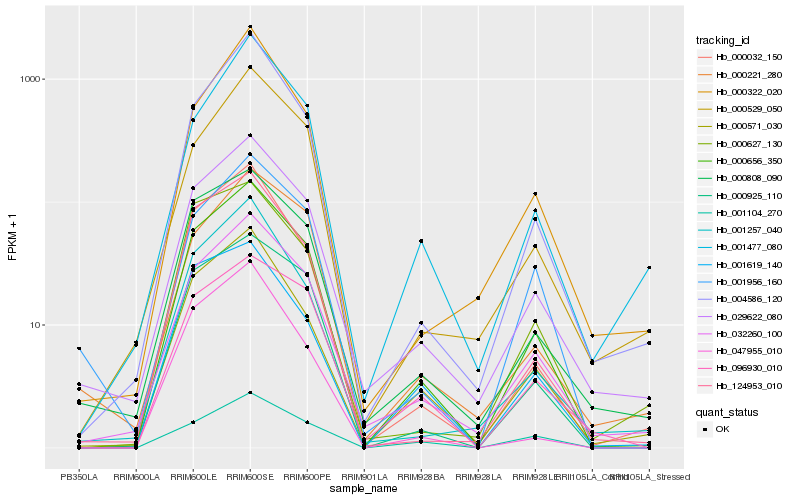
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_350 |
0.0 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_032260_100 |
0.0768776518 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 3 |
Hb_029622_080 |
0.0834327079 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 4 |
Hb_000925_110 |
0.085926537 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 5 |
Hb_000571_030 |
0.0924818423 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 6 |
Hb_000032_150 |
0.0960439635 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1691430 [Ricinus communis] |
| 7 |
Hb_000808_090 |
0.0987777544 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 8 |
Hb_124953_010 |
0.1007128233 |
- |
- |
hypothetical protein RCOM_1314600 [Ricinus communis] |
| 9 |
Hb_004586_120 |
0.1009230578 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 10 |
Hb_001257_040 |
0.1102141074 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 11 |
Hb_096930_010 |
0.1136903044 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 12 |
Hb_000529_050 |
0.1196182455 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 13 |
Hb_047955_010 |
0.1222577759 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 14 |
Hb_001104_270 |
0.122514793 |
- |
- |
hypothetical protein POPTR_0007s13980g [Populus trichocarpa] |
| 15 |
Hb_001956_160 |
0.1227222616 |
- |
- |
hypothetical protein L484_009438 [Morus notabilis] |
| 16 |
Hb_000627_130 |
0.1233057675 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 17 |
Hb_001619_140 |
0.1243310407 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 18 |
Hb_000322_020 |
0.1260591236 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 19 |
Hb_001477_080 |
0.1262545615 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 20 |
Hb_000221_280 |
0.126460225 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |