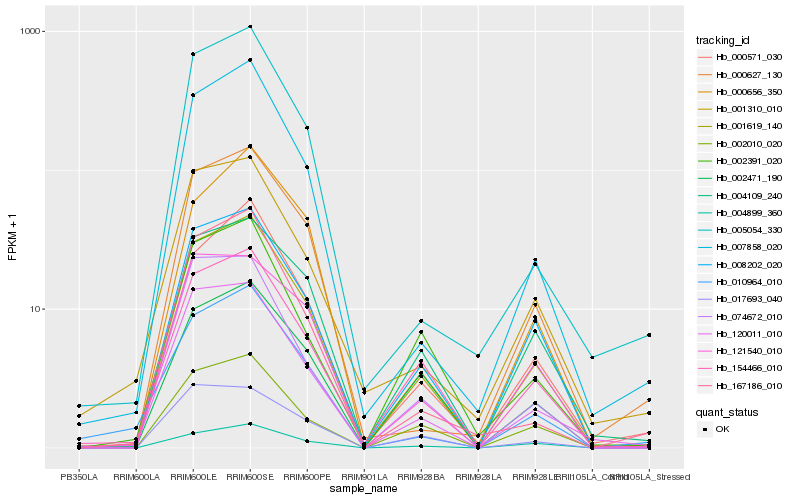
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_140 |
0.0 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_008202_020 |
0.0730950041 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 3 |
Hb_154466_010 |
0.0885021098 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 4 |
Hb_120011_010 |
0.0885216069 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 5 |
Hb_000571_030 |
0.093828831 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 6 |
Hb_004899_360 |
0.0972650089 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 7 |
Hb_007858_020 |
0.0978575537 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 8 |
Hb_004109_240 |
0.0982776599 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 9 |
Hb_002010_020 |
0.1002840783 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 10 |
Hb_000627_130 |
0.111412928 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 11 |
Hb_002471_190 |
0.113322212 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 12 |
Hb_002391_020 |
0.1180229285 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 13 |
Hb_005054_330 |
0.1192372356 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 14 |
Hb_010964_010 |
0.1214582173 |
- |
- |
- |
| 15 |
Hb_121540_010 |
0.121705498 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 16 |
Hb_074672_010 |
0.1218920267 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 17 |
Hb_167186_010 |
0.1230681027 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 18 |
Hb_001310_010 |
0.1232673911 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 19 |
Hb_017693_040 |
0.1241197501 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 20 |
Hb_000656_350 |
0.1243310407 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |