| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
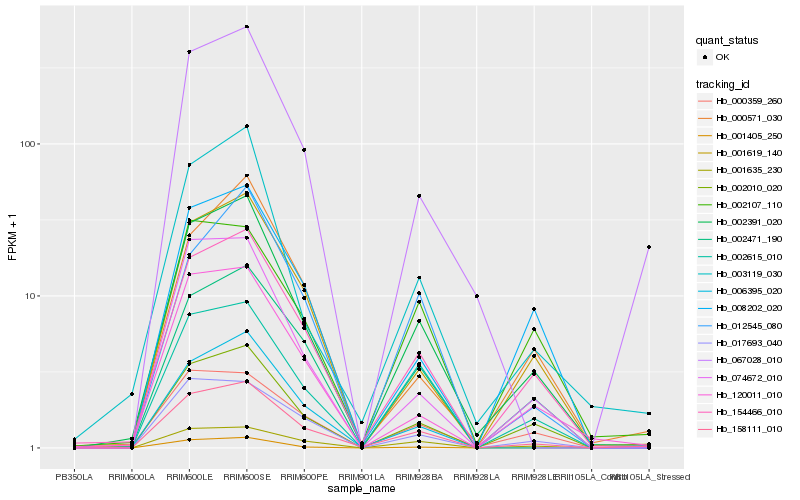
Hb_002391_020 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 2 |
Hb_154466_010 |
0.0775593658 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_002010_020 |
0.0915839271 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 4 |
Hb_158111_010 |
0.1030890512 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 5 |
Hb_003119_030 |
0.1092350414 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Jatropha curcas] |
| 6 |
Hb_002471_190 |
0.113976967 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 7 |
Hb_001619_140 |
0.1180229285 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_002615_010 |
0.1201124999 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 9 |
Hb_074672_010 |
0.1296931503 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 10 |
Hb_120011_010 |
0.1344403445 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_001635_230 |
0.1356130079 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_008202_020 |
0.1360490398 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_000359_260 |
0.1418185828 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 14 |
Hb_006395_020 |
0.1462700421 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 15 |
Hb_017693_040 |
0.1467126542 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 16 |
Hb_001405_250 |
0.1486095898 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 17 |
Hb_002107_110 |
0.1556506817 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000571_030 |
0.1562461731 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 19 |
Hb_012545_080 |
0.1585862766 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 20 |
Hb_067028_010 |
0.1605265488 |
- |
- |
BnaC03g72040D [Brassica napus] |