| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
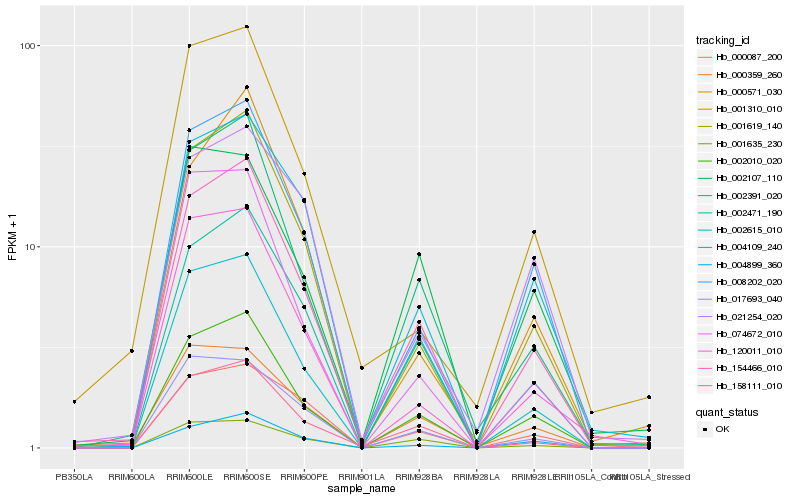
Hb_154466_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 2 |
Hb_002010_020 |
0.069018102 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 3 |
Hb_002471_190 |
0.0730569162 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 4 |
Hb_002391_020 |
0.0775593658 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 5 |
Hb_001619_140 |
0.0885021098 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_008202_020 |
0.0980216922 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 7 |
Hb_158111_010 |
0.1004388435 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 8 |
Hb_004109_240 |
0.1038034777 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 9 |
Hb_004899_360 |
0.1160311859 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 10 |
Hb_000359_260 |
0.1184719449 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 11 |
Hb_001635_230 |
0.1199961063 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_120011_010 |
0.1228061231 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 13 |
Hb_017693_040 |
0.125630942 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 14 |
Hb_000087_200 |
0.1272248411 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 15 |
Hb_002615_010 |
0.1281910063 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 16 |
Hb_000571_030 |
0.1333220129 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 17 |
Hb_001310_010 |
0.1439424045 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 18 |
Hb_002107_110 |
0.1445955261 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_074672_010 |
0.1458217963 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 20 |
Hb_021254_020 |
0.1494407524 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |