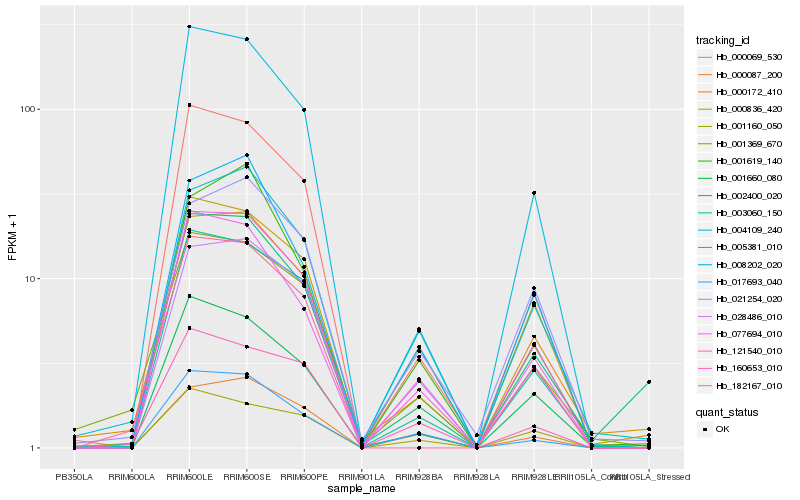
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_121540_010 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 2 |
Hb_003060_150 |
0.0655507621 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_005381_010 |
0.0700672424 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000069_530 |
0.0719608352 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 5 |
Hb_028486_010 |
0.0722401853 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 6 |
Hb_077694_010 |
0.0742076844 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 7 |
Hb_001660_080 |
0.0898701823 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 8 |
Hb_000836_420 |
0.0955777806 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_004109_240 |
0.0964029629 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 10 |
Hb_017693_040 |
0.0964044123 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 11 |
Hb_000172_410 |
0.0967436241 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_000087_200 |
0.0978775906 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 13 |
Hb_008202_020 |
0.107550358 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_001160_050 |
0.1118010404 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 15 |
Hb_182167_010 |
0.1162810824 |
- |
- |
hypothetical protein PHAVU_001G0486001g, partial [Phaseolus vulgaris] |
| 16 |
Hb_160653_010 |
0.1181149079 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 17 |
Hb_021254_020 |
0.1203340477 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 18 |
Hb_001369_670 |
0.1205649372 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 19 |
Hb_001619_140 |
0.121705498 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_002400_020 |
0.1235292157 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |