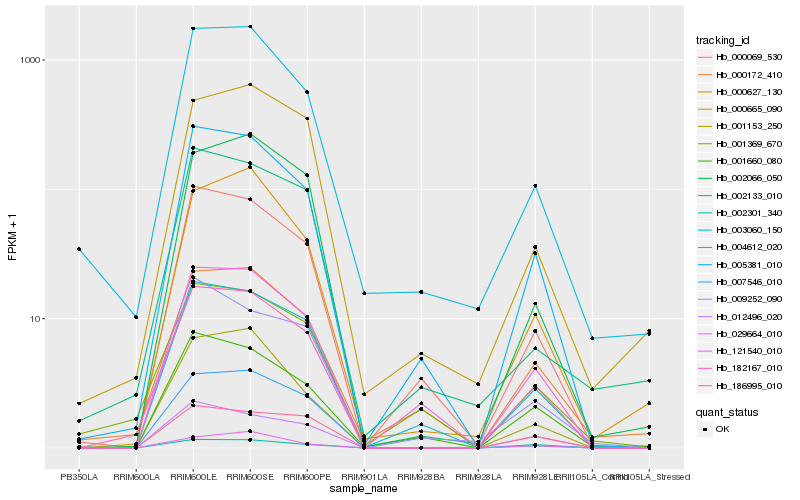
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182167_010 |
0.0 |
- |
- |
hypothetical protein PHAVU_001G0486001g, partial [Phaseolus vulgaris] |
| 2 |
Hb_005381_010 |
0.0795101025 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012496_020 |
0.1050956478 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 4 |
Hb_003060_150 |
0.1053403674 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000069_530 |
0.1074870816 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 6 |
Hb_121540_010 |
0.1162810824 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 7 |
Hb_001153_250 |
0.122851206 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 8 |
Hb_186995_010 |
0.1241428443 |
- |
- |
hypothetical protein POPTR_0012s01755g [Populus trichocarpa] |
| 9 |
Hb_000172_410 |
0.1276629477 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_004612_020 |
0.1288052585 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 11 |
Hb_002066_050 |
0.1313362562 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 12 |
Hb_001660_080 |
0.1327481129 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 13 |
Hb_001369_670 |
0.1357314307 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 14 |
Hb_000627_130 |
0.1372601943 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 15 |
Hb_000665_090 |
0.1404731763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_009252_090 |
0.1408783448 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_002301_340 |
0.1421535921 |
transcription factor |
TF Family: B3 |
leafy cotyledon 2 [Ricinus communis] |
| 18 |
Hb_029664_010 |
0.1436900368 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_007546_010 |
0.1465308058 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_002133_010 |
0.1465469916 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |