| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
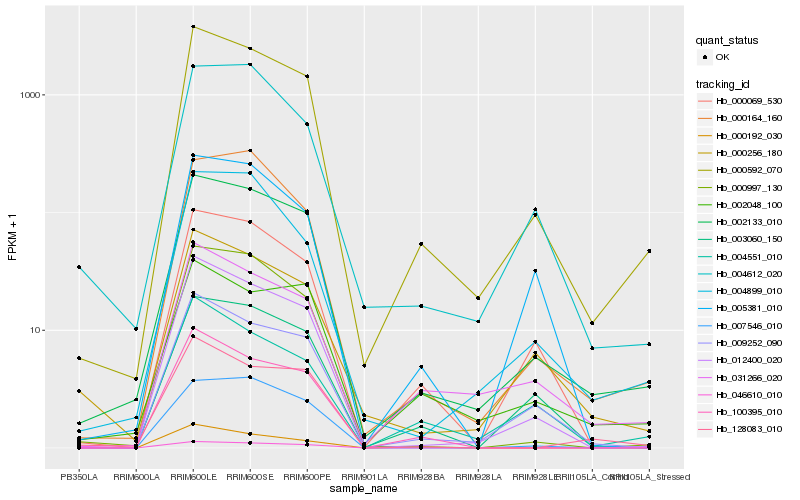
| 1 |
Hb_000592_070 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 2 |
Hb_002133_010 |
0.0572369323 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 3 |
Hb_012400_020 |
0.0797778036 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 4 |
Hb_009252_090 |
0.0962888959 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000069_530 |
0.0970993305 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 6 |
Hb_031266_020 |
0.1048715284 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003060_150 |
0.116991519 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000997_130 |
0.1175116006 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_100395_010 |
0.1203309202 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000256_180 |
0.1251330405 |
- |
- |
PREDICTED: uncharacterized protein LOC105637589 [Jatropha curcas] |
| 11 |
Hb_004551_010 |
0.1255418089 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 12 |
Hb_005381_010 |
0.1270221125 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000192_030 |
0.1280097249 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 14 |
Hb_046610_010 |
0.1301737513 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_004899_010 |
0.1317739952 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 16 |
Hb_004612_020 |
0.1331325411 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 17 |
Hb_000164_160 |
0.1336751153 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_128083_010 |
0.1372850657 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_002048_100 |
0.1384221116 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Jatropha curcas] |
| 20 |
Hb_007546_010 |
0.1395223313 |
- |
- |
unnamed protein product [Vitis vinifera] |