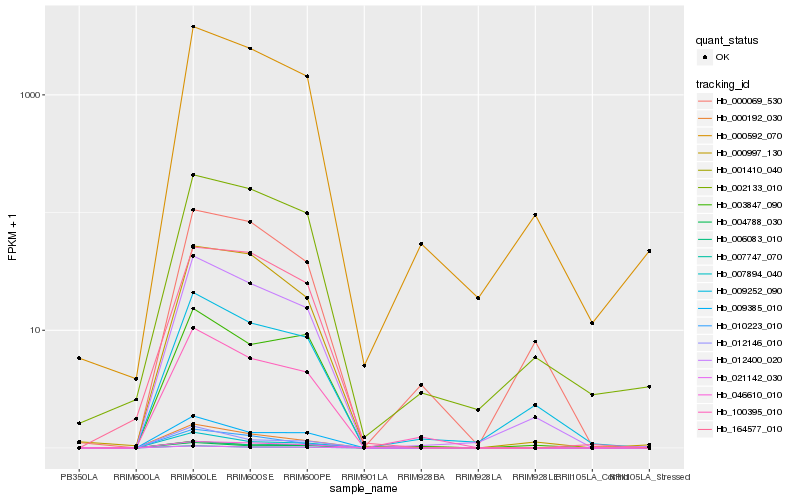
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012400_020 |
0.0 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 2 |
Hb_000592_070 |
0.0797778036 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 3 |
Hb_009252_090 |
0.0910582191 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_021142_030 |
0.0957377943 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 5 |
Hb_046610_010 |
0.103956108 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000997_130 |
0.1040460712 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_002133_010 |
0.1042965023 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 8 |
Hb_100395_010 |
0.1089082445 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_009385_010 |
0.1099786483 |
- |
- |
Cysteine/Histidine-rich C1 domain family protein, putative [Theobroma cacao] |
| 10 |
Hb_000069_530 |
0.1112984626 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 11 |
Hb_001410_040 |
0.1135295422 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 12 |
Hb_007747_070 |
0.1146634881 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |
| 13 |
Hb_007894_040 |
0.1150656195 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000192_030 |
0.1165366933 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 15 |
Hb_010223_010 |
0.1212875277 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 16 |
Hb_006083_010 |
0.1220306057 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_003847_090 |
0.1247494218 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 18 |
Hb_012146_010 |
0.128976136 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 19 |
Hb_004788_030 |
0.1303539838 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_164577_010 |
0.1332558647 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |