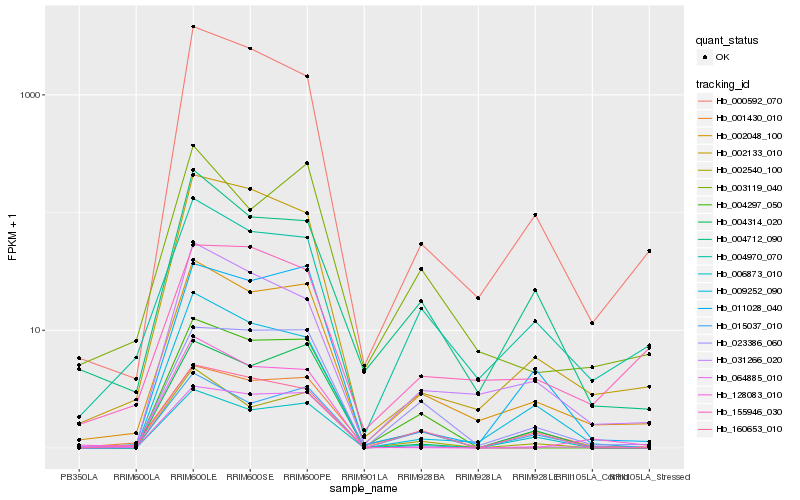
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002048_100 |
0.0 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Jatropha curcas] |
| 2 |
Hb_001430_010 |
0.1163747021 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004970_070 |
0.1268635462 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 4 |
Hb_002133_010 |
0.1305352139 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 5 |
Hb_031266_020 |
0.1309533084 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003119_040 |
0.1381118728 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 7 |
Hb_015037_010 |
0.1381441063 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 8 |
Hb_000592_070 |
0.1384221116 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 9 |
Hb_023386_060 |
0.1407794127 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 10 |
Hb_004712_090 |
0.14155882 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 11 |
Hb_002540_100 |
0.142637731 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 12 |
Hb_011028_040 |
0.1452463595 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_006873_010 |
0.1461605621 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_009252_090 |
0.1467151662 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_160653_010 |
0.1488894881 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 16 |
Hb_004297_050 |
0.1554819571 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_004314_020 |
0.1569744086 |
- |
- |
Auxin-responsive GH3 family protein [Theobroma cacao] |
| 18 |
Hb_128083_010 |
0.1577613379 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_064885_010 |
0.1611895252 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 20 |
Hb_155946_030 |
0.1621647755 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |