| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
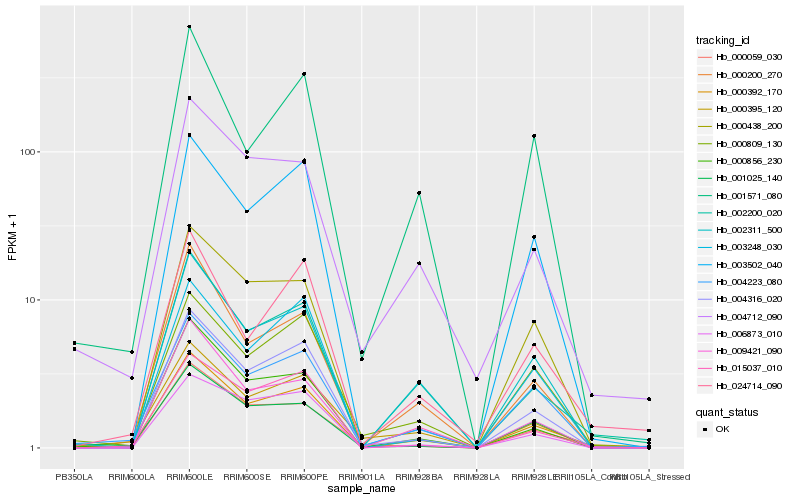
Hb_000392_170 |
0.0 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 2 |
Hb_004316_020 |
0.0962152882 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 3 |
Hb_002311_500 |
0.1004899636 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 4 |
Hb_002200_020 |
0.1023461408 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein RCOM_0137680 [Ricinus communis] |
| 5 |
Hb_004223_080 |
0.1060682748 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000856_230 |
0.106832111 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 7 |
Hb_000200_270 |
0.107055343 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000059_030 |
0.1167909717 |
- |
- |
PREDICTED: chaperone protein dnaJ 10 [Jatropha curcas] |
| 9 |
Hb_015037_010 |
0.123878319 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 10 |
Hb_009421_090 |
0.1239843381 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 11 |
Hb_000809_130 |
0.1239874723 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 12 |
Hb_001025_140 |
0.1282217848 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 13 |
Hb_006873_010 |
0.1302004457 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_024714_090 |
0.1302105316 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 15 |
Hb_004712_090 |
0.1307072012 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 16 |
Hb_003248_030 |
0.1307289978 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_003502_040 |
0.1323441418 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 18 |
Hb_000395_120 |
0.1324169201 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 19 |
Hb_001571_080 |
0.1332689586 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 20 |
Hb_000438_200 |
0.134876468 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |