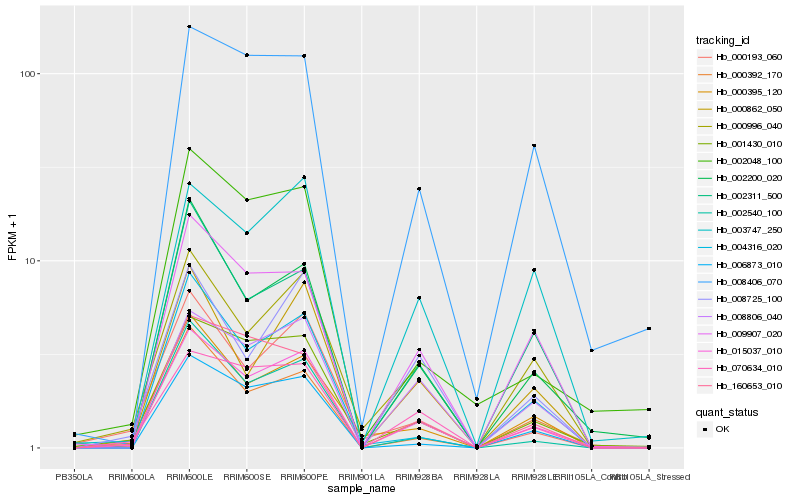
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015037_010 |
0.0 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 2 |
Hb_001430_010 |
0.1091750412 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002200_020 |
0.1202060555 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein RCOM_0137680 [Ricinus communis] |
| 4 |
Hb_070634_010 |
0.1225610376 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 5 |
Hb_000392_170 |
0.123878319 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 6 |
Hb_008806_040 |
0.1239710966 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 7 |
Hb_006873_010 |
0.1256883813 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_009907_020 |
0.1314346235 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 9 |
Hb_160653_010 |
0.1316127697 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 10 |
Hb_002540_100 |
0.1317433499 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 11 |
Hb_000996_040 |
0.1381152666 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 12 |
Hb_002048_100 |
0.1381441063 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Jatropha curcas] |
| 13 |
Hb_004316_020 |
0.1381450723 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 14 |
Hb_000395_120 |
0.1397290719 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 15 |
Hb_000193_060 |
0.1427594854 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 16 |
Hb_008406_070 |
0.144600867 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 17 |
Hb_008725_100 |
0.1447811427 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 18 |
Hb_002311_500 |
0.1448581147 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 19 |
Hb_003747_250 |
0.1477499977 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 20 |
Hb_000862_050 |
0.1482749041 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |