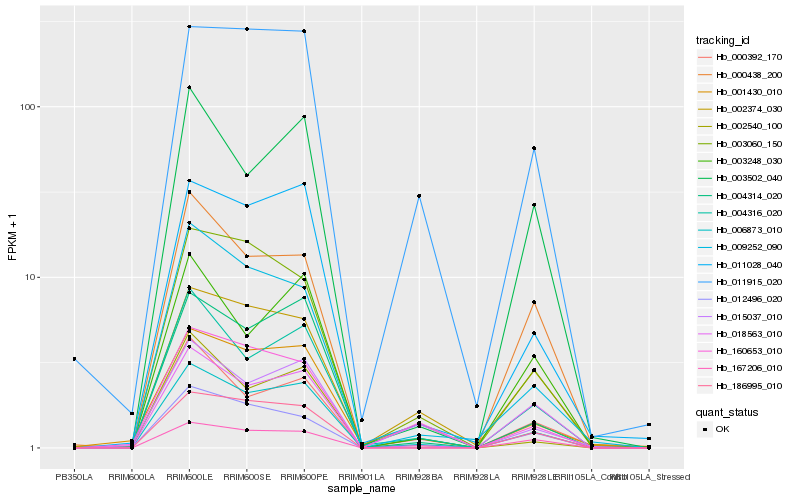
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006873_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 2 |
Hb_004316_020 |
0.0898510798 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 3 |
Hb_004314_020 |
0.0913419335 |
- |
- |
Auxin-responsive GH3 family protein [Theobroma cacao] |
| 4 |
Hb_011028_040 |
0.0952870473 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 5 |
Hb_018563_010 |
0.1133363654 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 6 |
Hb_186995_010 |
0.1159556212 |
- |
- |
hypothetical protein POPTR_0012s01755g [Populus trichocarpa] |
| 7 |
Hb_009252_090 |
0.1174732301 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_002540_100 |
0.1251167248 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 9 |
Hb_015037_010 |
0.1256883813 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 10 |
Hb_003060_150 |
0.12688109 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003502_040 |
0.1269142843 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 12 |
Hb_000392_170 |
0.1302004457 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 13 |
Hb_003248_030 |
0.1307826807 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_002374_030 |
0.132632962 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 15 |
Hb_167206_010 |
0.133316847 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 16 |
Hb_160653_010 |
0.1343020735 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 17 |
Hb_011915_020 |
0.1356285727 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 6, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_001430_010 |
0.1362411864 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000438_200 |
0.1366987855 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 20 |
Hb_012496_020 |
0.1385736519 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |