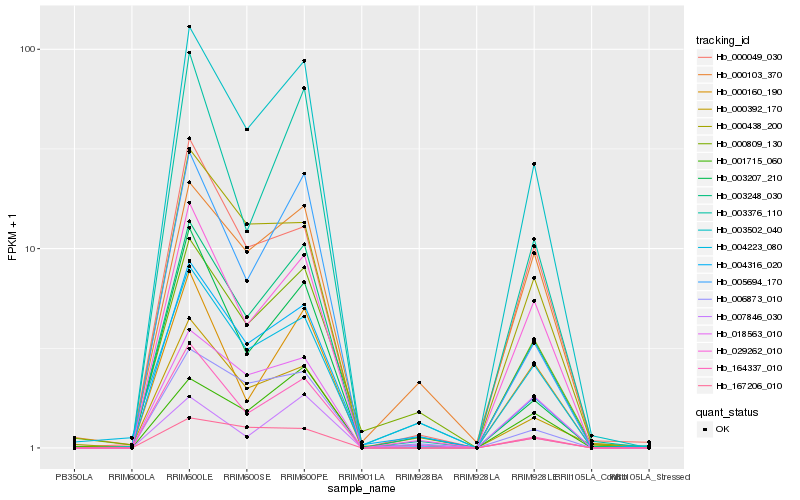
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003502_040 |
0.0 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 2 |
Hb_003248_030 |
0.0438311529 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_018563_010 |
0.0842620315 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 4 |
Hb_029262_010 |
0.0940722335 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 5 |
Hb_004316_020 |
0.0962512763 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 6 |
Hb_000438_200 |
0.1153731036 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 7 |
Hb_005694_170 |
0.1167527858 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 8 |
Hb_006873_010 |
0.1269142843 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_000809_130 |
0.1269578708 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 10 |
Hb_164337_010 |
0.1269791785 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 11 |
Hb_003376_110 |
0.1283892804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003207_210 |
0.1313377855 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 13 |
Hb_000160_190 |
0.1321937645 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 14 |
Hb_000392_170 |
0.1323441418 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 15 |
Hb_000049_030 |
0.1375532008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001715_060 |
0.139438356 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 17 |
Hb_167206_010 |
0.1404956837 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 18 |
Hb_000103_370 |
0.1416527633 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 19 |
Hb_007846_030 |
0.1425190351 |
- |
- |
Uncharacterized protein TCM_021839 [Theobroma cacao] |
| 20 |
Hb_004223_080 |
0.1433515408 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |