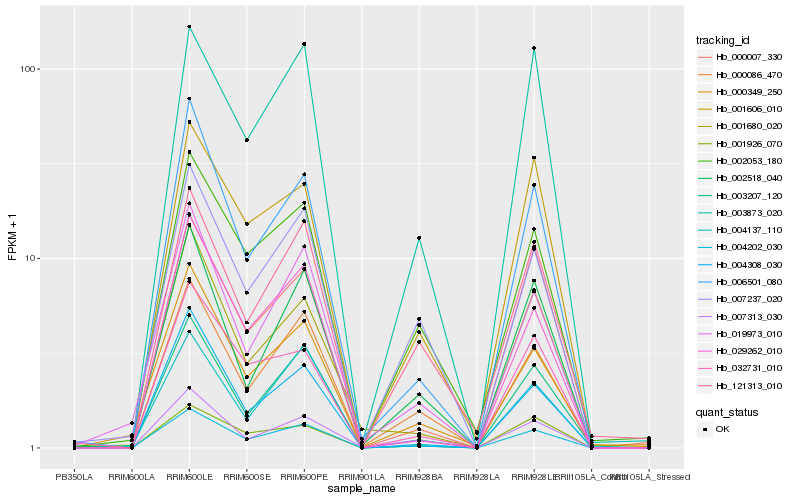
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004202_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 2 |
Hb_003207_120 |
0.0711972979 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 3 |
Hb_000086_470 |
0.0719199355 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 4 |
Hb_006501_080 |
0.0759213031 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 5 |
Hb_029262_010 |
0.0875363374 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 6 |
Hb_019973_010 |
0.0991832348 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004308_030 |
0.1017851088 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 8 |
Hb_002518_040 |
0.101841633 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 9 |
Hb_007237_020 |
0.1029151734 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 10 |
Hb_002053_180 |
0.1034782565 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 11 |
Hb_007313_030 |
0.1083761382 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 12 |
Hb_032731_010 |
0.1085999942 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |
| 13 |
Hb_000007_330 |
0.1093976848 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 14 |
Hb_004137_110 |
0.109866247 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |
| 15 |
Hb_000349_250 |
0.1099553246 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 16 |
Hb_121313_010 |
0.1103853767 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 17 |
Hb_003873_020 |
0.1105395957 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 18 |
Hb_001606_010 |
0.1120500772 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_001926_070 |
0.1135699935 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 20 |
Hb_001680_020 |
0.1152995157 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |