| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
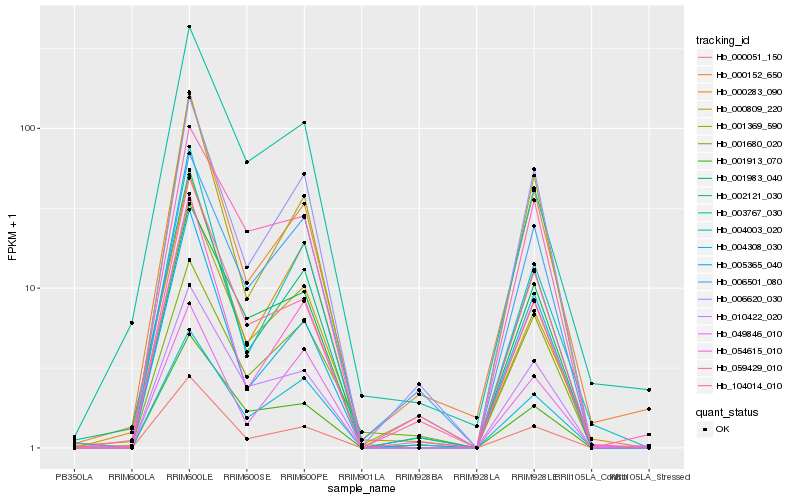
Hb_000809_220 |
0.0 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |
| 2 |
Hb_002121_030 |
0.072845873 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 3 |
Hb_000051_150 |
0.0732034918 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |
| 4 |
Hb_004308_030 |
0.0890701736 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 5 |
Hb_001913_070 |
0.0906790951 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 6 |
Hb_001983_040 |
0.0908965918 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 7 |
Hb_010422_020 |
0.0944251136 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 8 |
Hb_000152_650 |
0.0972038779 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_104014_010 |
0.1015267282 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 10 |
Hb_006620_030 |
0.1023383207 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 11 |
Hb_006501_080 |
0.1049587535 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 12 |
Hb_001369_590 |
0.1069849621 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 13 |
Hb_004003_020 |
0.1076371239 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000283_090 |
0.1114450938 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_054615_010 |
0.1180749597 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 16 |
Hb_059429_010 |
0.1190492689 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 17 |
Hb_005365_040 |
0.1205929997 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 18 |
Hb_003767_030 |
0.1217981747 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 19 |
Hb_001680_020 |
0.1226018015 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 20 |
Hb_049846_010 |
0.1227325287 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |