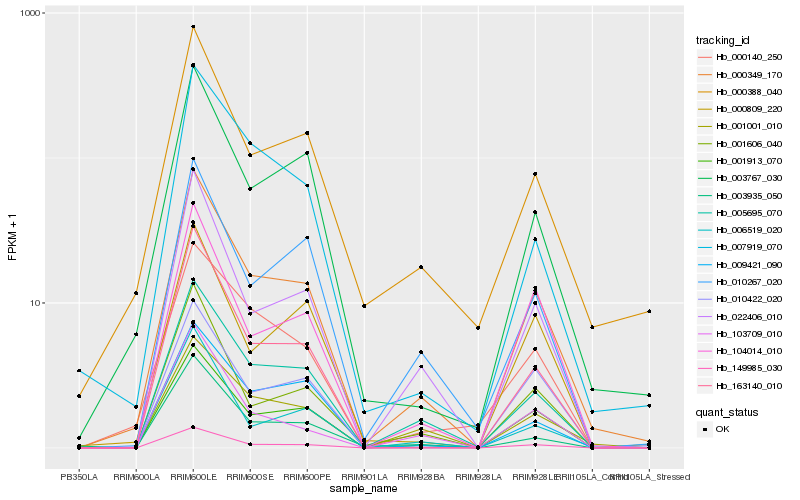
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000349_170 |
0.0 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_149985_030 |
0.0714528562 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 3 |
Hb_005695_070 |
0.0887617841 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 4 |
Hb_001913_070 |
0.0925209148 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 5 |
Hb_022406_010 |
0.1094464508 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_003767_030 |
0.111841233 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 7 |
Hb_000388_040 |
0.1124832975 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 8 |
Hb_163140_010 |
0.1157320094 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 9 |
Hb_001606_040 |
0.1186515921 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 10 |
Hb_001001_010 |
0.1211271874 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 11 |
Hb_104014_010 |
0.1244879828 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 12 |
Hb_010267_020 |
0.1313249104 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 13 |
Hb_006519_020 |
0.1324508495 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 14 |
Hb_003935_050 |
0.1330221494 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_000140_250 |
0.1331185288 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 16 |
Hb_007919_070 |
0.1354708306 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 17 |
Hb_009421_090 |
0.1376658863 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 18 |
Hb_010422_020 |
0.1393258927 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 19 |
Hb_000809_220 |
0.1398254022 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |
| 20 |
Hb_103709_010 |
0.1405526774 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |