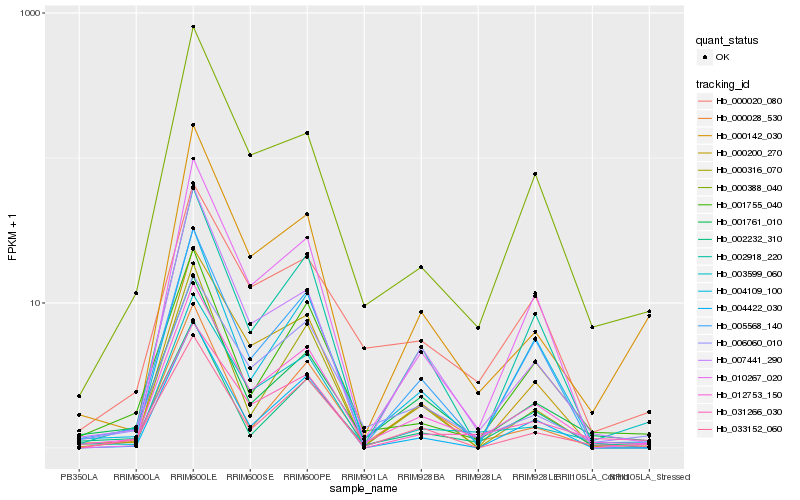
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012753_150 |
0.0 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 2 |
Hb_001761_010 |
0.1007809102 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 3 |
Hb_006060_010 |
0.1058466832 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 4 |
Hb_000388_040 |
0.1083431734 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 5 |
Hb_005568_140 |
0.1084362894 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_004109_100 |
0.1097550616 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_010267_020 |
0.1099633434 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 8 |
Hb_031266_030 |
0.1112018545 |
- |
- |
Kinase superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_002918_220 |
0.1113627191 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 10 |
Hb_007441_290 |
0.1115654423 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 11 |
Hb_000200_270 |
0.1145457656 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_033152_060 |
0.1206642419 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 [Jatropha curcas] |
| 13 |
Hb_003599_060 |
0.1255189043 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 14 |
Hb_001755_040 |
0.1279806867 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002232_310 |
0.1292894987 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 16 |
Hb_000028_530 |
0.1313213627 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 17 |
Hb_000142_030 |
0.1320190277 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000020_080 |
0.1332060174 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 19 |
Hb_004422_030 |
0.1349723069 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 20 |
Hb_000316_070 |
0.1353146772 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |