| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
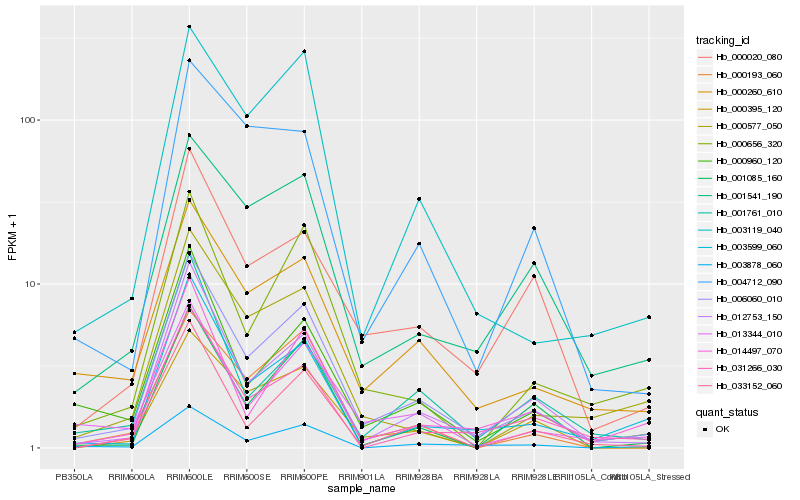
Hb_006060_010 |
0.0 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 2 |
Hb_012753_150 |
0.1058466832 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 3 |
Hb_000260_610 |
0.1236973623 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 4 |
Hb_003878_060 |
0.1289026267 |
- |
- |
hypothetical protein B456_007G326900, partial [Gossypium raimondii] |
| 5 |
Hb_000960_120 |
0.1292872272 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 6 |
Hb_003119_040 |
0.1303989361 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 7 |
Hb_013344_010 |
0.132494117 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 8 |
Hb_003599_060 |
0.1328667873 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_000656_320 |
0.1361792384 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 10 |
Hb_000193_060 |
0.1410806031 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 11 |
Hb_000395_120 |
0.1413727193 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 12 |
Hb_033152_060 |
0.1436147218 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 [Jatropha curcas] |
| 13 |
Hb_001541_190 |
0.1449424445 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 14 |
Hb_031266_030 |
0.1458050482 |
- |
- |
Kinase superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000577_050 |
0.1461581925 |
- |
- |
- |
| 16 |
Hb_014497_070 |
0.1468352589 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 17 |
Hb_000020_080 |
0.1482851412 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 18 |
Hb_001085_160 |
0.1485180991 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 19 |
Hb_001761_010 |
0.1509191486 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 20 |
Hb_004712_090 |
0.1512096357 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |