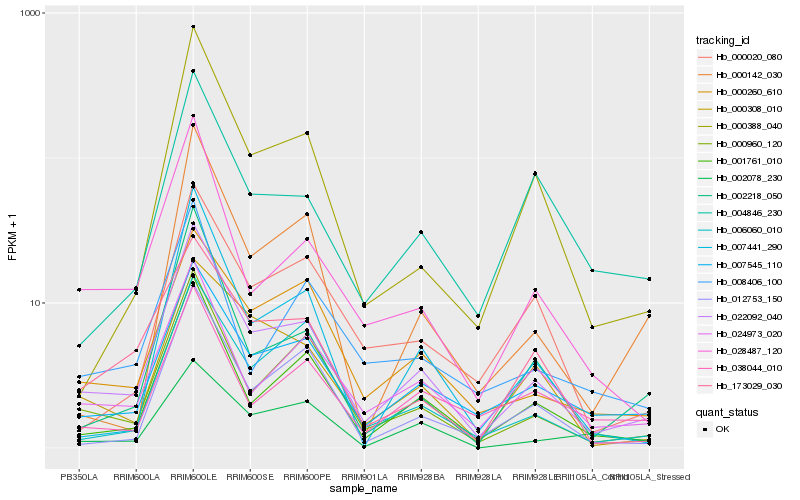
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022092_040 |
0.0 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 2 |
Hb_000960_120 |
0.1163200382 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 3 |
Hb_007545_110 |
0.1176766611 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_001761_010 |
0.1224308978 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 5 |
Hb_028487_120 |
0.1436032682 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 6 |
Hb_000260_610 |
0.1479787759 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 7 |
Hb_002218_050 |
0.1483401412 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 8 |
Hb_024973_020 |
0.1490085378 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 9 |
Hb_173029_030 |
0.1533788643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012753_150 |
0.1540981779 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 11 |
Hb_000388_040 |
0.1543482376 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 12 |
Hb_000308_010 |
0.1569867354 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 13 |
Hb_007441_290 |
0.160532312 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 14 |
Hb_006060_010 |
0.1658043375 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 15 |
Hb_000142_030 |
0.1694330556 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_008406_100 |
0.1733150724 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000020_080 |
0.1747295765 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 18 |
Hb_002078_230 |
0.1797162372 |
- |
- |
PREDICTED: HORMA domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_004846_230 |
0.1804551239 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_038044_010 |
0.1807417949 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |