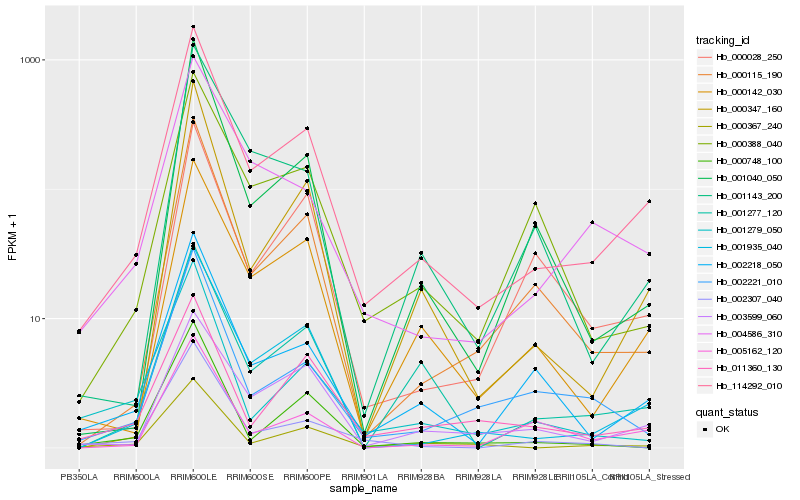
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114292_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001935_040 |
0.1102376854 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 3 |
Hb_000142_030 |
0.1270341287 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002218_050 |
0.1319305757 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 5 |
Hb_000115_190 |
0.1372626736 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 6 |
Hb_000347_160 |
0.1377418297 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 7 |
Hb_000367_240 |
0.1435929492 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |
| 8 |
Hb_004586_310 |
0.1438721164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003599_060 |
0.1491598969 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 10 |
Hb_001040_050 |
0.1541490704 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 11 |
Hb_001277_120 |
0.1563616944 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 12 |
Hb_001143_200 |
0.165669467 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 13 |
Hb_002221_010 |
0.1675667457 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |
| 14 |
Hb_011360_130 |
0.1679420813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000388_040 |
0.1680799053 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 16 |
Hb_005162_120 |
0.1683249385 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 17 |
Hb_001279_050 |
0.1705957372 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 18 |
Hb_002307_040 |
0.1719733934 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000748_100 |
0.1735296596 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 20 |
Hb_000028_250 |
0.1759986804 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |