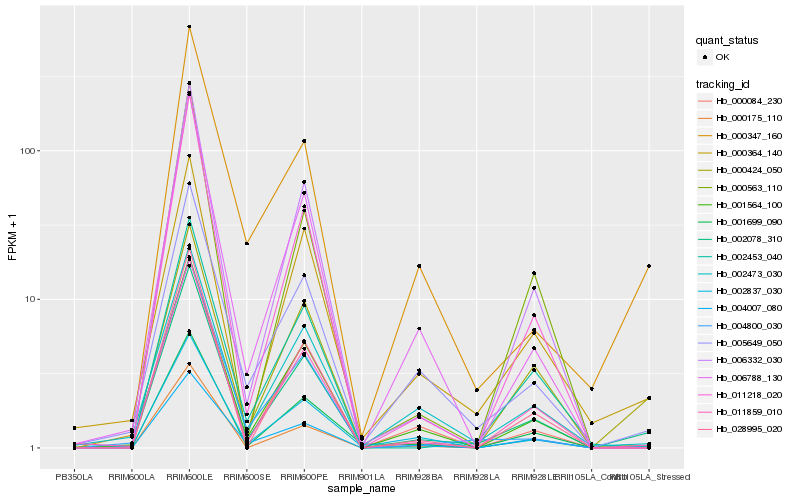
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_310 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 2 |
Hb_011859_010 |
0.0886080536 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 3 |
Hb_001699_090 |
0.0978031377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001564_100 |
0.1003701396 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 5 |
Hb_006332_030 |
0.1039819766 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 6 |
Hb_011218_020 |
0.1072964583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005649_050 |
0.1093063812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_028995_020 |
0.1108835665 |
- |
- |
PREDICTED: uncharacterized protein LOC105642143 [Jatropha curcas] |
| 9 |
Hb_000424_050 |
0.1110013555 |
- |
- |
erecta, putative [Ricinus communis] |
| 10 |
Hb_000084_230 |
0.1124705827 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 11 |
Hb_006788_130 |
0.1125054463 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 12 |
Hb_002473_030 |
0.1142120564 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 13 |
Hb_000364_140 |
0.1165867742 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 14 |
Hb_000347_160 |
0.1205326921 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 15 |
Hb_000175_110 |
0.1213759992 |
- |
- |
hypothetical protein JCGZ_06125 [Jatropha curcas] |
| 16 |
Hb_002453_040 |
0.122822837 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 17 |
Hb_004800_030 |
0.1233512876 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 18 |
Hb_000563_110 |
0.1250762806 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 19 |
Hb_002837_030 |
0.1264547925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004007_080 |
0.1272982775 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |