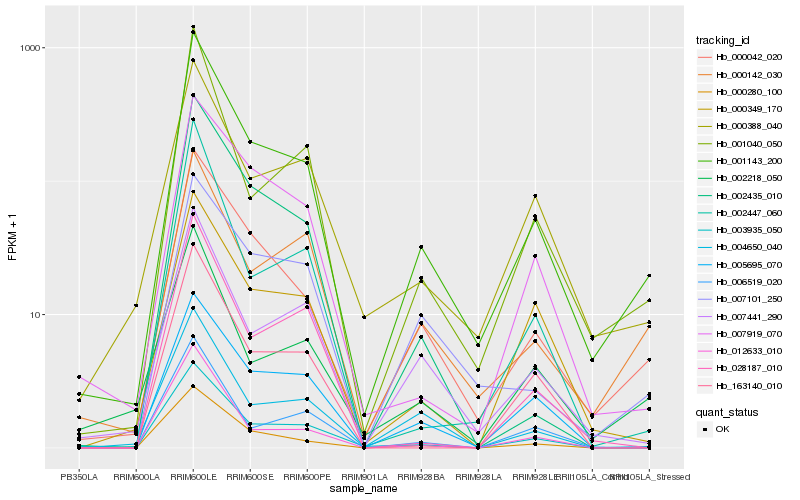
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001143_200 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 2 |
Hb_000042_020 |
0.0884574335 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000280_100 |
0.1094880964 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 4 |
Hb_003935_050 |
0.1187008266 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_001040_050 |
0.1267547542 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 6 |
Hb_007441_290 |
0.1329224857 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 7 |
Hb_000388_040 |
0.1347305511 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 8 |
Hb_012633_010 |
0.1353773038 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 9 |
Hb_004650_040 |
0.1364826298 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 10 |
Hb_005695_070 |
0.1389204113 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 11 |
Hb_000142_030 |
0.1392692573 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_007919_070 |
0.1415499153 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 13 |
Hb_002218_050 |
0.1429342767 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 14 |
Hb_002447_060 |
0.1468140061 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006519_020 |
0.1477774503 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_000349_170 |
0.1477931265 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_163140_010 |
0.1508526081 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 18 |
Hb_002435_010 |
0.1515846386 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 19 |
Hb_028187_010 |
0.151816641 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 20 |
Hb_007101_250 |
0.1521013314 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |