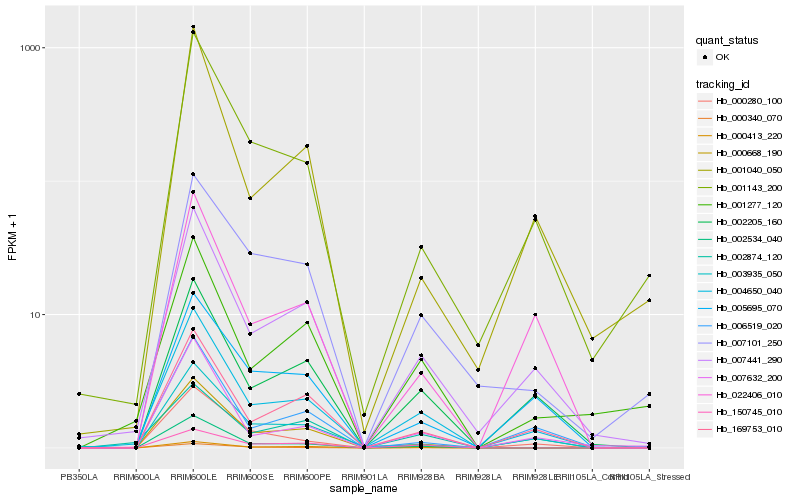
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004650_040 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 2 |
Hb_007441_290 |
0.0910223764 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 3 |
Hb_002205_160 |
0.1180373662 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 4 |
Hb_169753_010 |
0.1214389179 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 5 |
Hb_007632_200 |
0.135447558 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 6 |
Hb_001143_200 |
0.1364826298 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 7 |
Hb_022406_010 |
0.1473109255 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_001277_120 |
0.1479729185 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 9 |
Hb_002534_040 |
0.1481503529 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 10 |
Hb_003935_050 |
0.1546213388 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000413_220 |
0.1555869916 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 12 |
Hb_005695_070 |
0.1565548872 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 13 |
Hb_006519_020 |
0.1565709834 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 14 |
Hb_000280_100 |
0.1584885425 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 15 |
Hb_000668_190 |
0.1591653029 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 16 |
Hb_150745_010 |
0.1614320892 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 17 |
Hb_000340_070 |
0.1619743546 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 18 |
Hb_001040_050 |
0.1621937302 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 19 |
Hb_002874_120 |
0.1630597872 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 20 |
Hb_007101_250 |
0.1647295635 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |