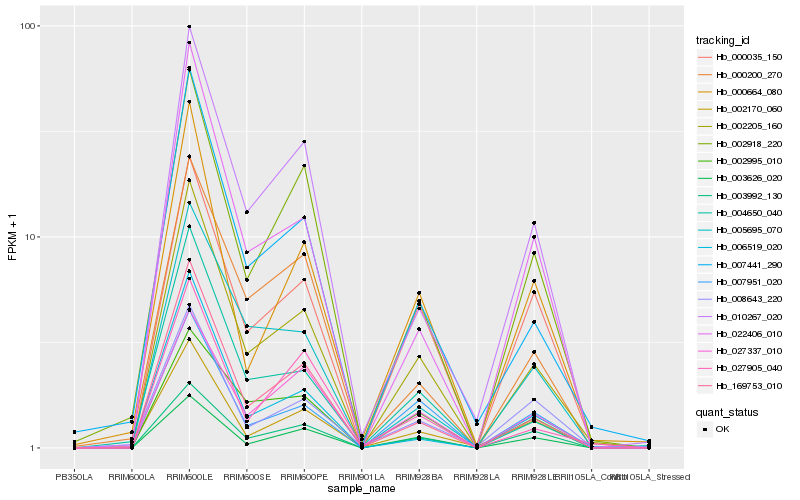
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_160 |
0.0 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 2 |
Hb_169753_010 |
0.0886546911 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 3 |
Hb_007441_290 |
0.0960596063 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 4 |
Hb_000035_150 |
0.1062055824 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 5 |
Hb_003992_130 |
0.1063331515 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_007951_020 |
0.1114234406 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 7 |
Hb_022406_010 |
0.1128073075 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002918_220 |
0.1163550848 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 9 |
Hb_010267_020 |
0.1175449196 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 10 |
Hb_004650_040 |
0.1180373662 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 11 |
Hb_027337_010 |
0.1222543847 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 12 |
Hb_000664_080 |
0.1231952269 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X2 [Jatropha curcas] |
| 13 |
Hb_003626_020 |
0.1238194696 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 14 |
Hb_005695_070 |
0.123914686 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 15 |
Hb_002170_060 |
0.1258742892 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_002995_010 |
0.1307519744 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 17 |
Hb_006519_020 |
0.1310585239 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_000200_270 |
0.1321319192 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_027905_040 |
0.1335790124 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: E2F transcription factor-like E2FF [Jatropha curcas] |
| 20 |
Hb_008643_220 |
0.1366742589 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |