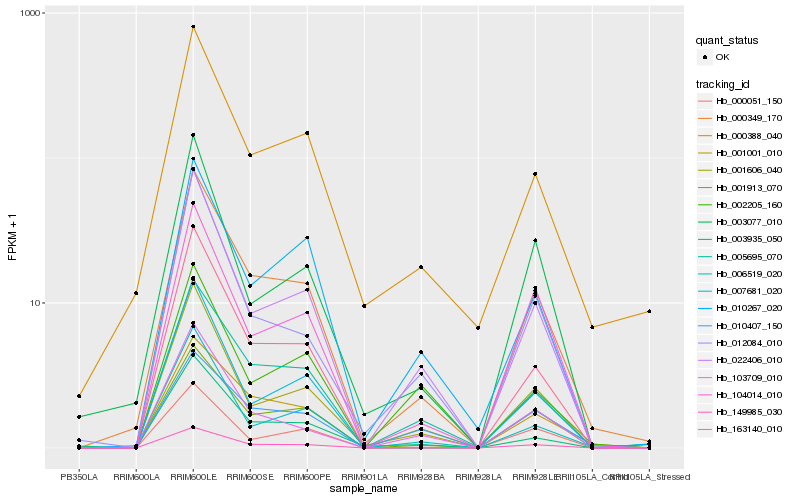
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_149985_030 |
0.0 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 2 |
Hb_022406_010 |
0.065025359 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000349_170 |
0.0714528562 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 4 |
Hb_005695_070 |
0.0763477471 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 5 |
Hb_001913_070 |
0.085845886 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 6 |
Hb_001606_040 |
0.0949314896 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 7 |
Hb_103709_010 |
0.1014857415 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 8 |
Hb_104014_010 |
0.1096200767 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 9 |
Hb_163140_010 |
0.1118303456 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 10 |
Hb_012084_010 |
0.1128963611 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 11 |
Hb_006519_020 |
0.1141553251 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_001001_010 |
0.1202672713 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 13 |
Hb_010267_020 |
0.1205405356 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 14 |
Hb_003935_050 |
0.1266124149 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_007681_020 |
0.1309827394 |
- |
- |
transferase, putative [Ricinus communis] |
| 16 |
Hb_010407_150 |
0.1336999991 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 17 |
Hb_003077_010 |
0.1380518227 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_002205_160 |
0.1411655391 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 19 |
Hb_000051_150 |
0.1430156288 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |
| 20 |
Hb_000388_040 |
0.1441331885 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |