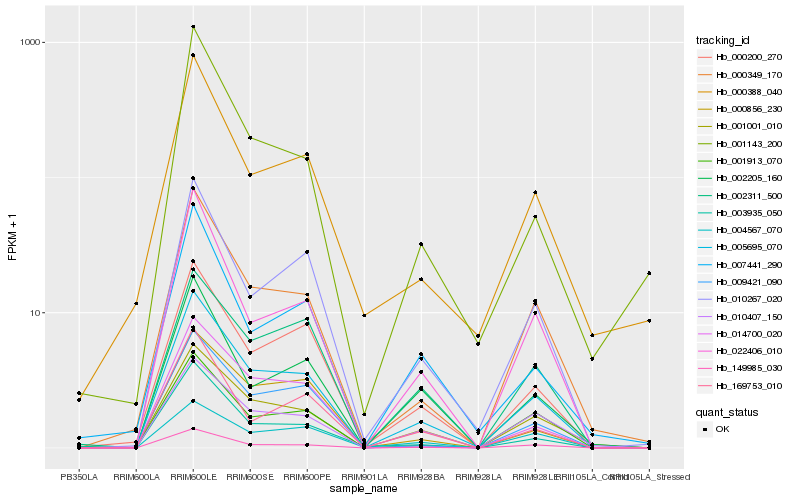
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005695_070 |
0.0 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 2 |
Hb_149985_030 |
0.0763477471 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 3 |
Hb_001001_010 |
0.0864923531 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 4 |
Hb_000349_170 |
0.0887617841 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 5 |
Hb_010267_020 |
0.0990975268 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 6 |
Hb_000200_270 |
0.1050546291 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_022406_010 |
0.1093750646 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_001913_070 |
0.1157246785 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 9 |
Hb_009421_090 |
0.1176250934 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 10 |
Hb_010407_150 |
0.1199423246 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 11 |
Hb_003935_050 |
0.1199444563 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_002205_160 |
0.123914686 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 13 |
Hb_169753_010 |
0.1319624467 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 14 |
Hb_002311_500 |
0.1333476228 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 15 |
Hb_000856_230 |
0.1349267866 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 16 |
Hb_000388_040 |
0.1362015184 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 17 |
Hb_004567_070 |
0.1364129355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007441_290 |
0.1364440308 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 19 |
Hb_014700_020 |
0.1374103734 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 20 |
Hb_001143_200 |
0.1389204113 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |