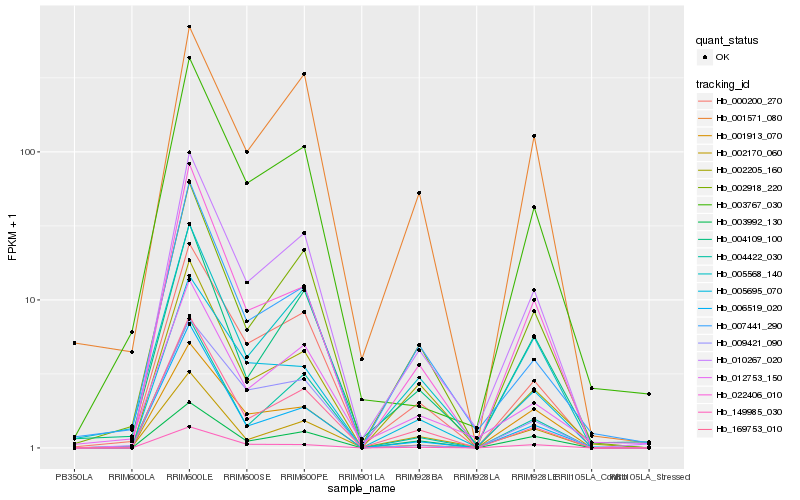
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010267_020 |
0.0 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 2 |
Hb_000200_270 |
0.0775392186 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_169753_010 |
0.0892837615 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 4 |
Hb_002918_220 |
0.0911018389 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 5 |
Hb_005568_140 |
0.0950833071 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_004422_030 |
0.0974733046 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 7 |
Hb_005695_070 |
0.0990975268 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 8 |
Hb_012753_150 |
0.1099633434 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 9 |
Hb_004109_100 |
0.1106973565 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 10 |
Hb_022406_010 |
0.111565342 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_002205_160 |
0.1175449196 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 12 |
Hb_149985_030 |
0.1205405356 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 13 |
Hb_003767_030 |
0.1215332361 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 14 |
Hb_001913_070 |
0.1219114087 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 15 |
Hb_003992_130 |
0.1222686506 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_006519_020 |
0.1232408195 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 17 |
Hb_009421_090 |
0.1234429821 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 18 |
Hb_007441_290 |
0.1247651635 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 19 |
Hb_001571_080 |
0.1247844356 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 20 |
Hb_002170_060 |
0.1257705167 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |