| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
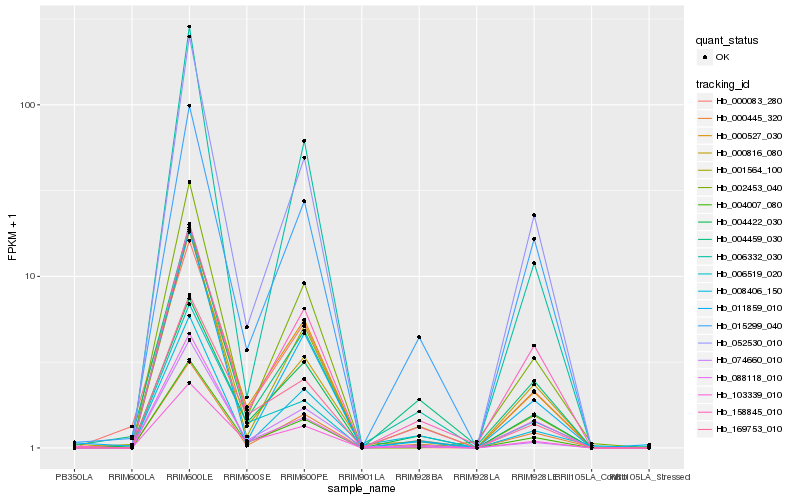
Hb_004007_080 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 2 |
Hb_000527_030 |
0.0666767909 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 3 |
Hb_006519_020 |
0.0839170215 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 4 |
Hb_052530_010 |
0.092681793 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000816_080 |
0.0927033086 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 6 |
Hb_088118_010 |
0.0932592938 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 7 |
Hb_103339_010 |
0.0952802519 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_001564_100 |
0.0953171151 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 9 |
Hb_004459_030 |
0.0977642377 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004422_030 |
0.0995491822 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 11 |
Hb_169753_010 |
0.1007620381 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 12 |
Hb_002453_040 |
0.1017286744 |
- |
- |
PREDICTED: uncharacterized protein LOC105636487 [Jatropha curcas] |
| 13 |
Hb_074660_010 |
0.1024988788 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 14 |
Hb_158845_010 |
0.1046980482 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 [Jatropha curcas] |
| 15 |
Hb_011859_010 |
0.1067743505 |
- |
- |
hypothetical protein VITISV_027923 [Vitis vinifera] |
| 16 |
Hb_015299_040 |
0.1094706388 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 17 |
Hb_000083_280 |
0.1098573825 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_008406_150 |
0.1105514298 |
- |
- |
PREDICTED: mitotic checkpoint serine/threonine-protein kinase BUB1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000445_320 |
0.112103835 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_006332_030 |
0.1139004363 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |