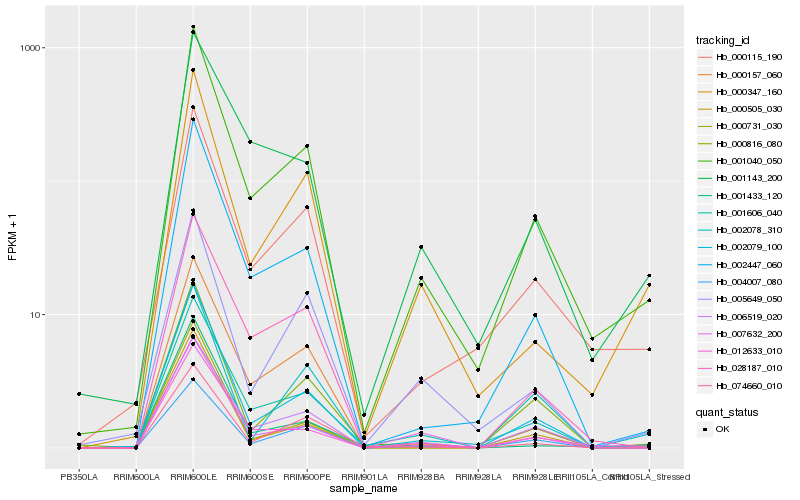
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001040_050 |
0.0 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 2 |
Hb_002447_060 |
0.0843118547 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000115_190 |
0.0870484705 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 4 |
Hb_006519_020 |
0.0927545407 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_002079_100 |
0.1004204261 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 6 |
Hb_012633_010 |
0.1068863986 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 7 |
Hb_000347_160 |
0.1100131772 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 8 |
Hb_007632_200 |
0.112720323 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 9 |
Hb_000157_060 |
0.1144733255 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004007_080 |
0.1195094352 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 11 |
Hb_028187_010 |
0.1200973254 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 12 |
Hb_001143_200 |
0.1267547542 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 13 |
Hb_000731_030 |
0.1291838758 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000816_080 |
0.1306128632 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 15 |
Hb_005649_050 |
0.1321686995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002078_310 |
0.1336859222 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 17 |
Hb_074660_010 |
0.1344763098 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 18 |
Hb_001433_120 |
0.1346025147 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 19 |
Hb_000505_030 |
0.1346054541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001606_040 |
0.1348021499 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |