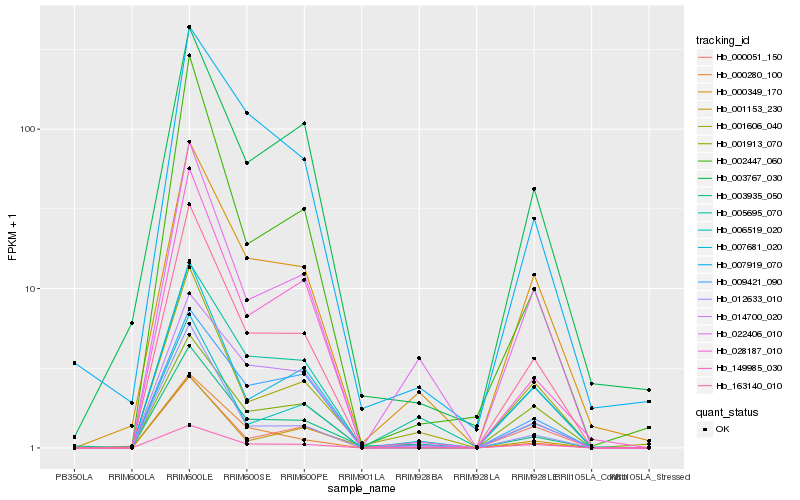
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163140_010 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 2 |
Hb_007681_020 |
0.0773634733 |
- |
- |
transferase, putative [Ricinus communis] |
| 3 |
Hb_028187_010 |
0.1055558537 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 4 |
Hb_149985_030 |
0.1118303456 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 5 |
Hb_012633_010 |
0.1147850802 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 6 |
Hb_006519_020 |
0.114890029 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_000349_170 |
0.1157320094 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 8 |
Hb_002447_060 |
0.1184363215 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003935_050 |
0.1205498466 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_022406_010 |
0.1226345713 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003767_030 |
0.1284661826 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 12 |
Hb_001606_040 |
0.1308067594 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 13 |
Hb_001153_230 |
0.1325663959 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 14 |
Hb_001913_070 |
0.1356750778 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 15 |
Hb_009421_090 |
0.1387144396 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 16 |
Hb_000051_150 |
0.1391038079 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |
| 17 |
Hb_014700_020 |
0.1405812899 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 18 |
Hb_005695_070 |
0.1413728409 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 19 |
Hb_007919_070 |
0.1414935723 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 20 |
Hb_000280_100 |
0.1434401301 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |