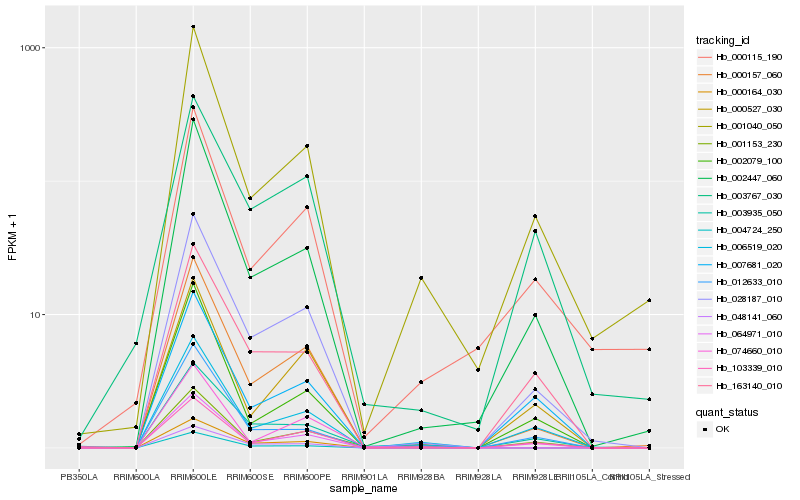
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028187_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 2 |
Hb_000157_060 |
0.062487451 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002447_060 |
0.0979992651 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_163140_010 |
0.1055558537 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 5 |
Hb_006519_020 |
0.1076566065 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_001153_230 |
0.1136522108 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 7 |
Hb_000527_030 |
0.1180257053 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 8 |
Hb_007681_020 |
0.1189274344 |
- |
- |
transferase, putative [Ricinus communis] |
| 9 |
Hb_000164_030 |
0.1193359396 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 10 |
Hb_001040_050 |
0.1200973254 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 11 |
Hb_003935_050 |
0.1263126229 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_074660_010 |
0.1264548474 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 13 |
Hb_012633_010 |
0.1272141291 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 14 |
Hb_064971_010 |
0.1274253402 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 15 |
Hb_003767_030 |
0.1277719186 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 16 |
Hb_048141_060 |
0.1279910537 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 17 |
Hb_002079_100 |
0.129487383 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 18 |
Hb_004724_250 |
0.1311061741 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 19 |
Hb_103339_010 |
0.1338896287 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000115_190 |
0.1353087225 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |