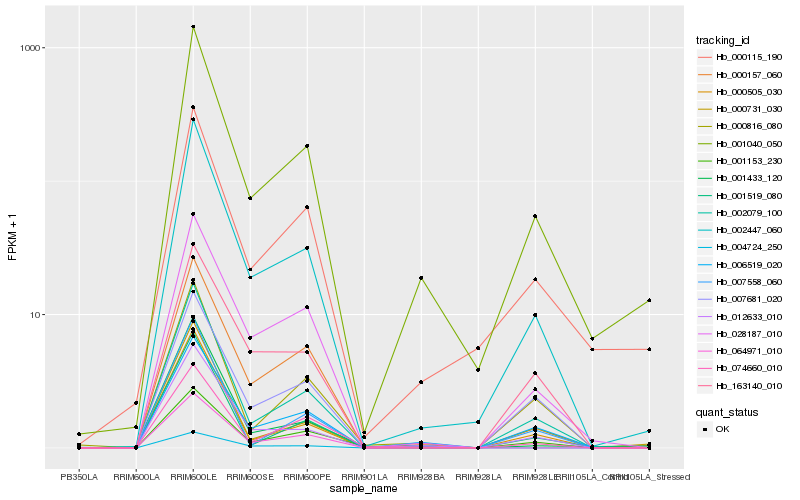
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002447_060 |
0.0 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002079_100 |
0.0693833431 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 3 |
Hb_012633_010 |
0.0732886847 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 4 |
Hb_001040_050 |
0.0843118547 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 5 |
Hb_000157_060 |
0.092288013 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_028187_010 |
0.0979992651 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 7 |
Hb_000505_030 |
0.0997941007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006519_020 |
0.1130079229 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_000731_030 |
0.1177945769 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_163140_010 |
0.1184363215 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 11 |
Hb_007681_020 |
0.1207756995 |
- |
- |
transferase, putative [Ricinus communis] |
| 12 |
Hb_001153_230 |
0.1228984362 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 13 |
Hb_001433_120 |
0.1262612498 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 14 |
Hb_000115_190 |
0.1306674178 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 15 |
Hb_064971_010 |
0.1349139681 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 16 |
Hb_001519_080 |
0.1357436769 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 17 |
Hb_004724_250 |
0.1357451814 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 18 |
Hb_000816_080 |
0.1357748439 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 19 |
Hb_074660_010 |
0.1362521856 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 20 |
Hb_007558_060 |
0.1406099384 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |