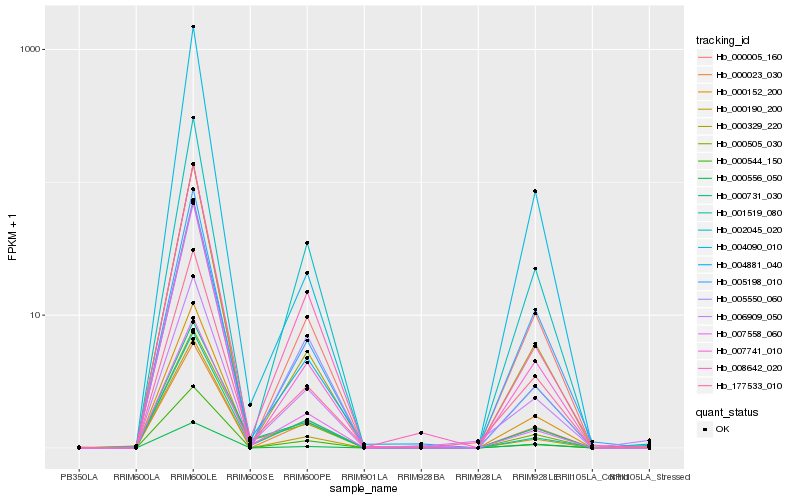
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_200 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000005_160 |
0.0526349557 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 3 |
Hb_177533_010 |
0.0561771246 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 4 |
Hb_007741_010 |
0.0577979203 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 5 |
Hb_000544_150 |
0.0769464785 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 6 |
Hb_000556_050 |
0.0774873488 |
- |
- |
- |
| 7 |
Hb_005198_010 |
0.0790007427 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 8 |
Hb_000329_220 |
0.0802634999 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 9 |
Hb_000505_030 |
0.0808647303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004881_040 |
0.0842294888 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 11 |
Hb_007558_060 |
0.0857239849 |
- |
- |
PREDICTED: uncharacterized protein LOC105644482 [Jatropha curcas] |
| 12 |
Hb_002045_020 |
0.0878351267 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 13 |
Hb_000023_030 |
0.0888405824 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 14 |
Hb_000731_030 |
0.0896599508 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000190_200 |
0.0928363913 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 16 |
Hb_008642_020 |
0.0974603547 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 17 |
Hb_005550_060 |
0.0985661804 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_006909_050 |
0.1012021412 |
transcription factor |
TF Family: OFP |
hypothetical protein POPTR_0008s01840g [Populus trichocarpa] |
| 19 |
Hb_004090_010 |
0.1018075341 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 20 |
Hb_001519_080 |
0.1034951621 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |