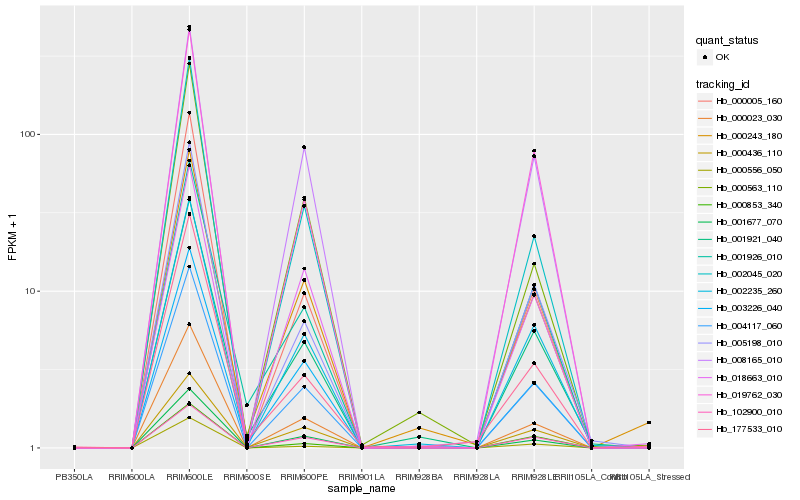
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_060 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g33811 [Jatropha curcas] |
| 2 |
Hb_002235_260 |
0.0279115315 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 3 |
Hb_000023_030 |
0.0455516243 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 4 |
Hb_001921_040 |
0.0588226863 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 5 |
Hb_008165_010 |
0.0632018363 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein [Solanum lycopersicum] |
| 6 |
Hb_002045_020 |
0.0633411772 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 7 |
Hb_019762_030 |
0.0640068513 |
- |
- |
hypothetical protein POPTR_0002s08720g [Populus trichocarpa] |
| 8 |
Hb_003226_040 |
0.0664126976 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 9 |
Hb_005198_010 |
0.0697170492 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 10 |
Hb_000243_180 |
0.0719433032 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1 [Jatropha curcas] |
| 11 |
Hb_001926_010 |
0.0789155249 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 12 |
Hb_000436_110 |
0.081392931 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 13 |
Hb_000005_160 |
0.0851055949 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 14 |
Hb_001677_070 |
0.087041001 |
- |
- |
PREDICTED: putative caffeoyl-CoA O-methyltransferase At1g67980 [Jatropha curcas] |
| 15 |
Hb_018663_010 |
0.0894797952 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 16 |
Hb_000853_340 |
0.0921399699 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |
| 17 |
Hb_000556_050 |
0.0922106181 |
- |
- |
- |
| 18 |
Hb_102900_010 |
0.0935416417 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |
| 19 |
Hb_177533_010 |
0.0948863281 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 20 |
Hb_000563_110 |
0.1006615313 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |