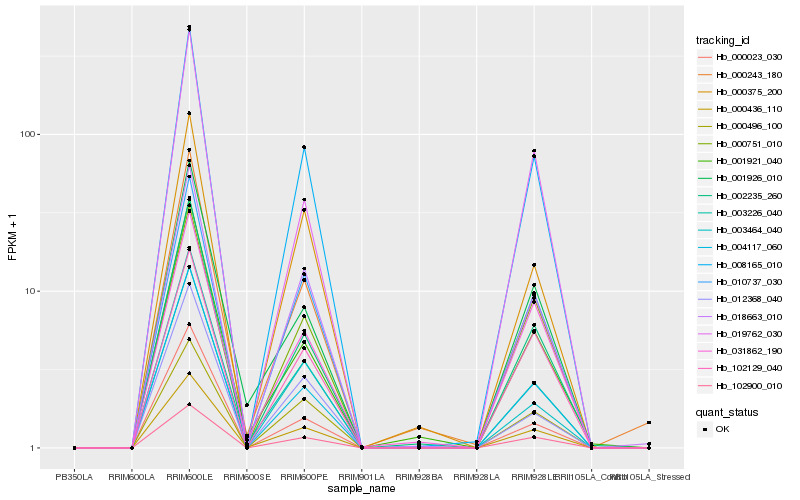
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008165_010 |
0.0 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein [Solanum lycopersicum] |
| 2 |
Hb_000436_110 |
0.0384168711 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 3 |
Hb_102900_010 |
0.0408693039 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |
| 4 |
Hb_018663_010 |
0.0415048946 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 5 |
Hb_010737_030 |
0.0460304251 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002235_260 |
0.0618499936 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 7 |
Hb_004117_060 |
0.0632018363 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g33811 [Jatropha curcas] |
| 8 |
Hb_000751_010 |
0.069976782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003226_040 |
0.0708246389 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 10 |
Hb_000496_100 |
0.0736169994 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 11 |
Hb_000375_200 |
0.0798472272 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 12 |
Hb_102129_040 |
0.0829277478 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000243_180 |
0.0829486081 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1 [Jatropha curcas] |
| 14 |
Hb_000023_030 |
0.0853680756 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 15 |
Hb_003464_040 |
0.0899999776 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 16 |
Hb_031862_190 |
0.0915841335 |
- |
- |
- |
| 17 |
Hb_012368_040 |
0.0930540357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_019762_030 |
0.0943928786 |
- |
- |
hypothetical protein POPTR_0002s08720g [Populus trichocarpa] |
| 19 |
Hb_001921_040 |
0.0950285719 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 20 |
Hb_001926_010 |
0.0952887007 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |