| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
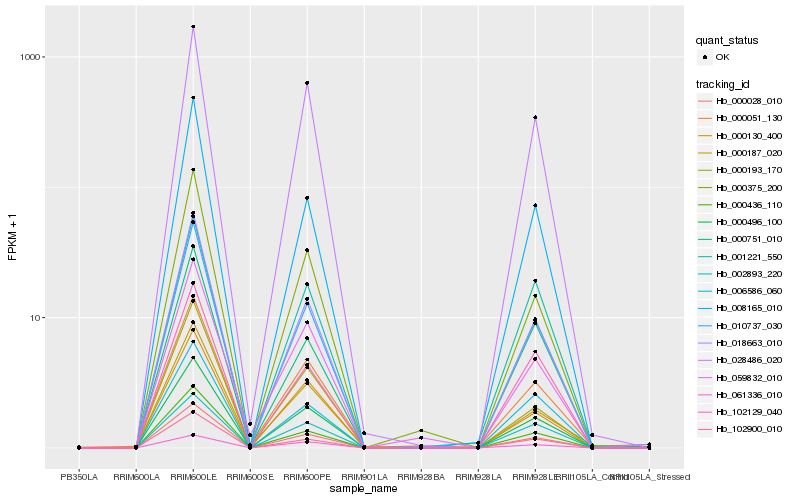
Hb_000496_100 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 2 |
Hb_010737_030 |
0.0349509995 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_028486_020 |
0.0558342778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_018663_010 |
0.05633385 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 5 |
Hb_102900_010 |
0.058227682 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000130_400 |
0.0685296167 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0001s38320g [Populus trichocarpa] |
| 7 |
Hb_008165_010 |
0.0736169994 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein [Solanum lycopersicum] |
| 8 |
Hb_000436_110 |
0.0740942337 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 9 |
Hb_000051_130 |
0.0776890507 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 10 |
Hb_000375_200 |
0.0792754428 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 11 |
Hb_102129_040 |
0.0824080117 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000751_010 |
0.0853785313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_059832_010 |
0.0857457681 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 14 |
Hb_006586_060 |
0.0858971125 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 15 |
Hb_001221_550 |
0.08727638 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000028_010 |
0.0878087305 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_061336_010 |
0.0886828751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000187_020 |
0.0925800513 |
- |
- |
Xanthine/uracil permease family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_000193_170 |
0.0937153029 |
transcription factor |
TF Family: MYB |
Myb domain 16-like protein [Theobroma cacao] |
| 20 |
Hb_002893_220 |
0.0948251783 |
- |
- |
hypothetical protein B456_001G004300 [Gossypium raimondii] |