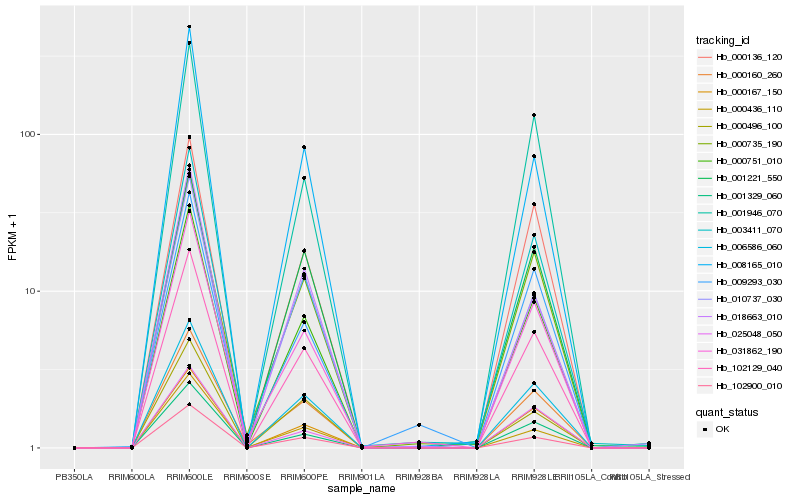
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000751_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_102129_040 |
0.024657581 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_102900_010 |
0.0375665654 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006586_060 |
0.0424753831 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 5 |
Hb_031862_190 |
0.0495182198 |
- |
- |
- |
| 6 |
Hb_001329_060 |
0.0499902385 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 7 |
Hb_003411_070 |
0.0532694666 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 8 |
Hb_000735_190 |
0.0544997337 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 9 |
Hb_000160_260 |
0.059877905 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 10 |
Hb_000436_110 |
0.0664240684 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 11 |
Hb_008165_010 |
0.069976782 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein [Solanum lycopersicum] |
| 12 |
Hb_010737_030 |
0.0703412337 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_000167_150 |
0.0707768008 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 14 |
Hb_000136_120 |
0.0748569876 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 15 |
Hb_001946_070 |
0.078659532 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000496_100 |
0.0853785313 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 17 |
Hb_001221_550 |
0.087526221 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_009293_030 |
0.088514993 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 19 |
Hb_025048_050 |
0.0895439854 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 20 |
Hb_018663_010 |
0.0910358821 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |