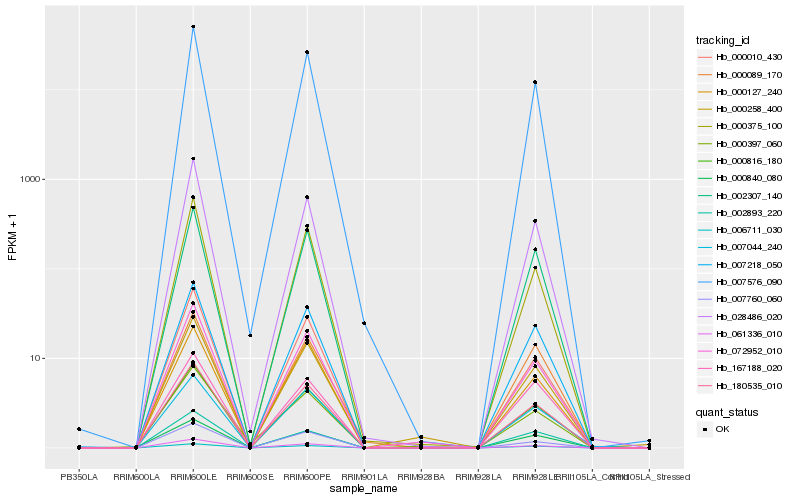
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 2 |
Hb_180535_010 |
0.0180433899 |
- |
- |
- |
| 3 |
Hb_061336_010 |
0.0196810362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007576_090 |
0.0301181783 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 5 |
Hb_007218_050 |
0.0340720149 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_002307_140 |
0.0440496525 |
transcription factor |
TF Family: AUX/IAA |
aux/IAA family protein [Populus trichocarpa] |
| 7 |
Hb_072952_010 |
0.0492699008 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 8 |
Hb_028486_020 |
0.0542994748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000258_400 |
0.0661677104 |
transcription factor |
TF Family: bHLH |
transcription regulator, putative [Ricinus communis] |
| 10 |
Hb_007760_060 |
0.0664727198 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17470g [Populus trichocarpa] |
| 11 |
Hb_006711_030 |
0.0684768125 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 12 |
Hb_000375_100 |
0.0691498761 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 13 |
Hb_002893_220 |
0.0712747496 |
- |
- |
hypothetical protein B456_001G004300 [Gossypium raimondii] |
| 14 |
Hb_000010_430 |
0.0751867539 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 15 |
Hb_000840_080 |
0.0762179438 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 16 |
Hb_007044_240 |
0.0771656881 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000127_240 |
0.0781910257 |
- |
- |
SAUR39 [Populus tomentosa] |
| 18 |
Hb_000089_170 |
0.0800928375 |
- |
- |
PREDICTED: uncharacterized protein LOC105134595 [Populus euphratica] |
| 19 |
Hb_167188_020 |
0.0827664298 |
- |
- |
PREDICTED: uncharacterized protein At1g05835 [Jatropha curcas] |
| 20 |
Hb_000397_060 |
0.0917710865 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |