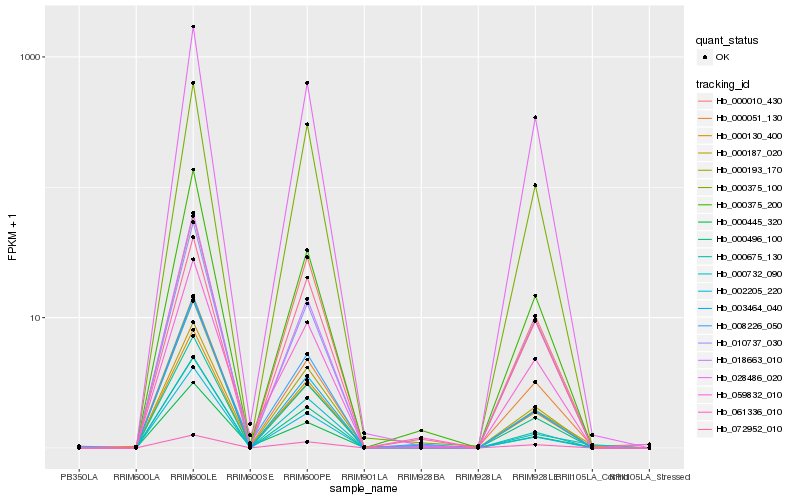
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_400 |
0.0 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0001s38320g [Populus trichocarpa] |
| 2 |
Hb_000375_200 |
0.0572482228 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 3 |
Hb_059832_010 |
0.0663692476 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 4 |
Hb_000496_100 |
0.0685296167 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 5 |
Hb_000193_170 |
0.0687778004 |
transcription factor |
TF Family: MYB |
Myb domain 16-like protein [Theobroma cacao] |
| 6 |
Hb_028486_020 |
0.0698676738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000187_020 |
0.0752947813 |
- |
- |
Xanthine/uracil permease family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_000010_430 |
0.0765634866 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 9 |
Hb_000375_100 |
0.0784475005 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 10 |
Hb_018663_010 |
0.0814863013 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 11 |
Hb_010737_030 |
0.0823043116 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_008226_050 |
0.0836201396 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 13 |
Hb_002205_220 |
0.0870710243 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 14 |
Hb_000051_130 |
0.0875384437 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 15 |
Hb_072952_010 |
0.0880924769 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 16 |
Hb_000675_130 |
0.092645248 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_061336_010 |
0.0948519042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000445_320 |
0.1005022406 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 19 |
Hb_000732_090 |
0.1014287487 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 20 |
Hb_003464_040 |
0.1024679306 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |