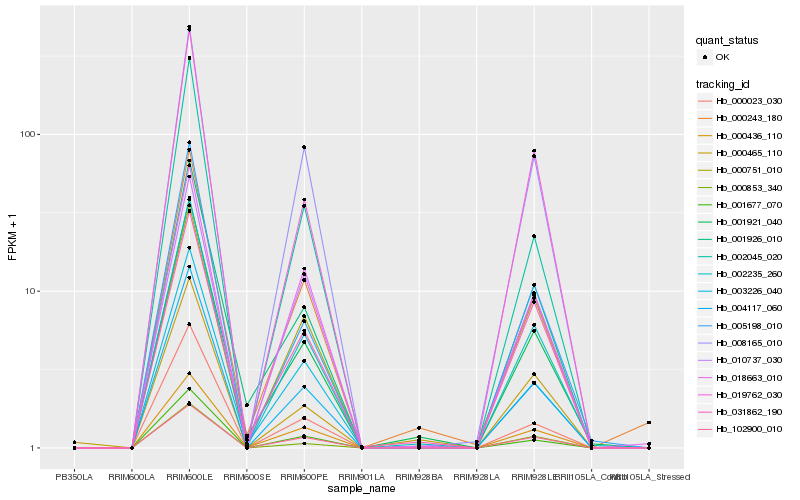
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_260 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 2 |
Hb_004117_060 |
0.0279115315 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g33811 [Jatropha curcas] |
| 3 |
Hb_001921_040 |
0.0464469663 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 4 |
Hb_019762_030 |
0.0618113622 |
- |
- |
hypothetical protein POPTR_0002s08720g [Populus trichocarpa] |
| 5 |
Hb_008165_010 |
0.0618499936 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein [Solanum lycopersicum] |
| 6 |
Hb_000023_030 |
0.0643707302 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 7 |
Hb_005198_010 |
0.0709530356 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 8 |
Hb_000243_180 |
0.0722571548 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1 [Jatropha curcas] |
| 9 |
Hb_003226_040 |
0.0732255358 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 10 |
Hb_001926_010 |
0.0736114859 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 11 |
Hb_000436_110 |
0.0748854149 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 12 |
Hb_002045_020 |
0.0797913422 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 13 |
Hb_102900_010 |
0.0869793828 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000853_340 |
0.0876254329 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |
| 15 |
Hb_018663_010 |
0.0893404628 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 16 |
Hb_001677_070 |
0.0921041348 |
- |
- |
PREDICTED: putative caffeoyl-CoA O-methyltransferase At1g67980 [Jatropha curcas] |
| 17 |
Hb_031862_190 |
0.0958297675 |
- |
- |
- |
| 18 |
Hb_000465_110 |
0.0975594349 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Jatropha curcas] |
| 19 |
Hb_000751_010 |
0.0983983685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010737_030 |
0.0992568044 |
- |
- |
unnamed protein product [Vitis vinifera] |