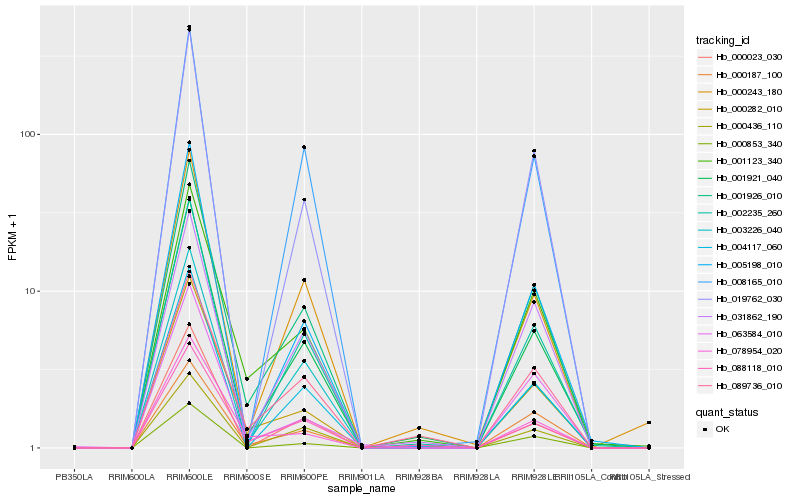
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001926_010 |
0.0 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 2 |
Hb_001921_040 |
0.0619943243 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 3 |
Hb_000282_010 |
0.0696818457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002235_260 |
0.0736114859 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 5 |
Hb_001123_340 |
0.07719323 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 6 |
Hb_019762_030 |
0.0775085825 |
- |
- |
hypothetical protein POPTR_0002s08720g [Populus trichocarpa] |
| 7 |
Hb_004117_060 |
0.0789155249 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g33811 [Jatropha curcas] |
| 8 |
Hb_000436_110 |
0.0815091114 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 9 |
Hb_088118_010 |
0.0852429045 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 10 |
Hb_000187_100 |
0.0874807623 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 11 |
Hb_003226_040 |
0.0891760144 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 12 |
Hb_031862_190 |
0.0921133184 |
- |
- |
- |
| 13 |
Hb_005198_010 |
0.0921819138 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 14 |
Hb_089736_010 |
0.0928043642 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 15 |
Hb_000853_340 |
0.0947031884 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |
| 16 |
Hb_008165_010 |
0.0952887007 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein [Solanum lycopersicum] |
| 17 |
Hb_000243_180 |
0.0956700415 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1 [Jatropha curcas] |
| 18 |
Hb_063584_010 |
0.0967876574 |
- |
- |
hypothetical protein JCGZ_10530 [Jatropha curcas] |
| 19 |
Hb_078954_020 |
0.0977139634 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 20 |
Hb_000023_030 |
0.1006587072 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |