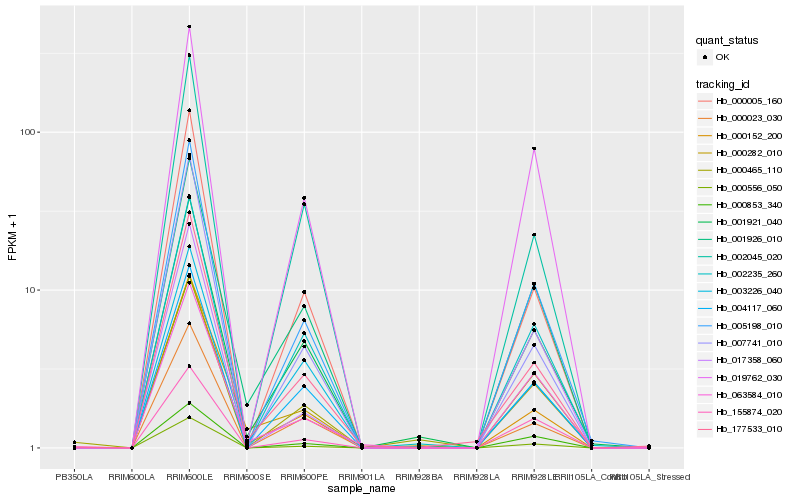
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005198_010 |
0.0 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 2 |
Hb_000556_050 |
0.0402203182 |
- |
- |
- |
| 3 |
Hb_019762_030 |
0.0611931389 |
- |
- |
hypothetical protein POPTR_0002s08720g [Populus trichocarpa] |
| 4 |
Hb_000005_160 |
0.0640709021 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 5 |
Hb_004117_060 |
0.0697170492 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g33811 [Jatropha curcas] |
| 6 |
Hb_002235_260 |
0.0709530356 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 7 |
Hb_177533_010 |
0.0741554766 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 8 |
Hb_001921_040 |
0.0748712622 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 9 |
Hb_000023_030 |
0.0762051464 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 10 |
Hb_000152_200 |
0.0790007427 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000853_340 |
0.084177748 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |
| 12 |
Hb_002045_020 |
0.0903817886 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 13 |
Hb_001926_010 |
0.0921819138 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 14 |
Hb_007741_010 |
0.0931456783 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 1 [Jatropha curcas] |
| 15 |
Hb_017358_060 |
0.0982577656 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 16 |
Hb_155874_020 |
0.1082320968 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 17 |
Hb_003226_040 |
0.1090030572 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 18 |
Hb_000465_110 |
0.1090746726 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Jatropha curcas] |
| 19 |
Hb_063584_010 |
0.1091357351 |
- |
- |
hypothetical protein JCGZ_10530 [Jatropha curcas] |
| 20 |
Hb_000282_010 |
0.1102466093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |