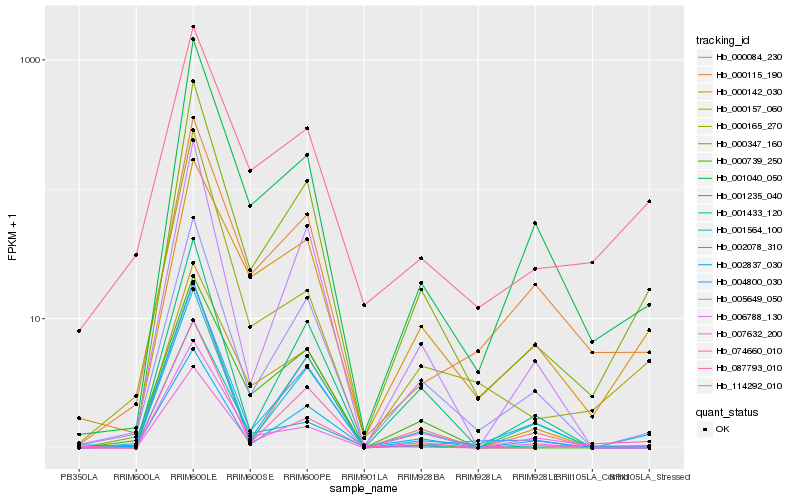
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_160 |
0.0 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 2 |
Hb_001040_050 |
0.1100131772 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 3 |
Hb_005649_050 |
0.1158534799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_087793_010 |
0.1191096714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002078_310 |
0.1205326921 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 6 |
Hb_006788_130 |
0.1338570762 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 7 |
Hb_114292_010 |
0.1377418297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004800_030 |
0.1387856929 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 9 |
Hb_000157_060 |
0.1404991121 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000115_190 |
0.1418314607 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 11 |
Hb_001235_040 |
0.1431656464 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 12 |
Hb_002837_030 |
0.1442083425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_074660_010 |
0.1450249804 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 14 |
Hb_000084_230 |
0.1475774749 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 15 |
Hb_000142_030 |
0.148946768 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_001564_100 |
0.1490693552 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 17 |
Hb_000165_270 |
0.1496517535 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 18 |
Hb_001433_120 |
0.1498591169 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 19 |
Hb_000739_250 |
0.1517071439 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 20 |
Hb_007632_200 |
0.1520834525 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |